
Page 30
Insights in Enzyme Research
ISSN: 2573-4466
E u r o S c i C o n C o n g r e s s o n
Enzymology and
Molecular Biology
A u g u s t 1 3 - 1 4 , 2 0 1 8
P a r i s , F r a n c e
Enzymology 2018
T
he globalenzyme market was estimated at $7,082 million as of 2017 and
is expected to reach $10,519 million in 2024. At a CAGR of 5.7% from 2018
to 2024, enzymes like transaminases are going to contribute the maximum for
this growth. Enzymes are molecular machineries used in various industries
such as pharmaceuticals, cosmetics; food and animal feed, paper and leather
processing, biofuel etc. Nevertheless, this has been possible only by the
breath-taking efforts of the chemists and biologists to evolve/engineer these
mysterious biomolecules to work the needful. The methodologies for this
research include the well-established directed evolution, rational redesign and
relatively less established yet much faster and accurate insilico methods. Main
agenda of an enzyme engineering project is to derive screening and selection
tools to obtain focused libraries of enzyme variants with desired qualities.
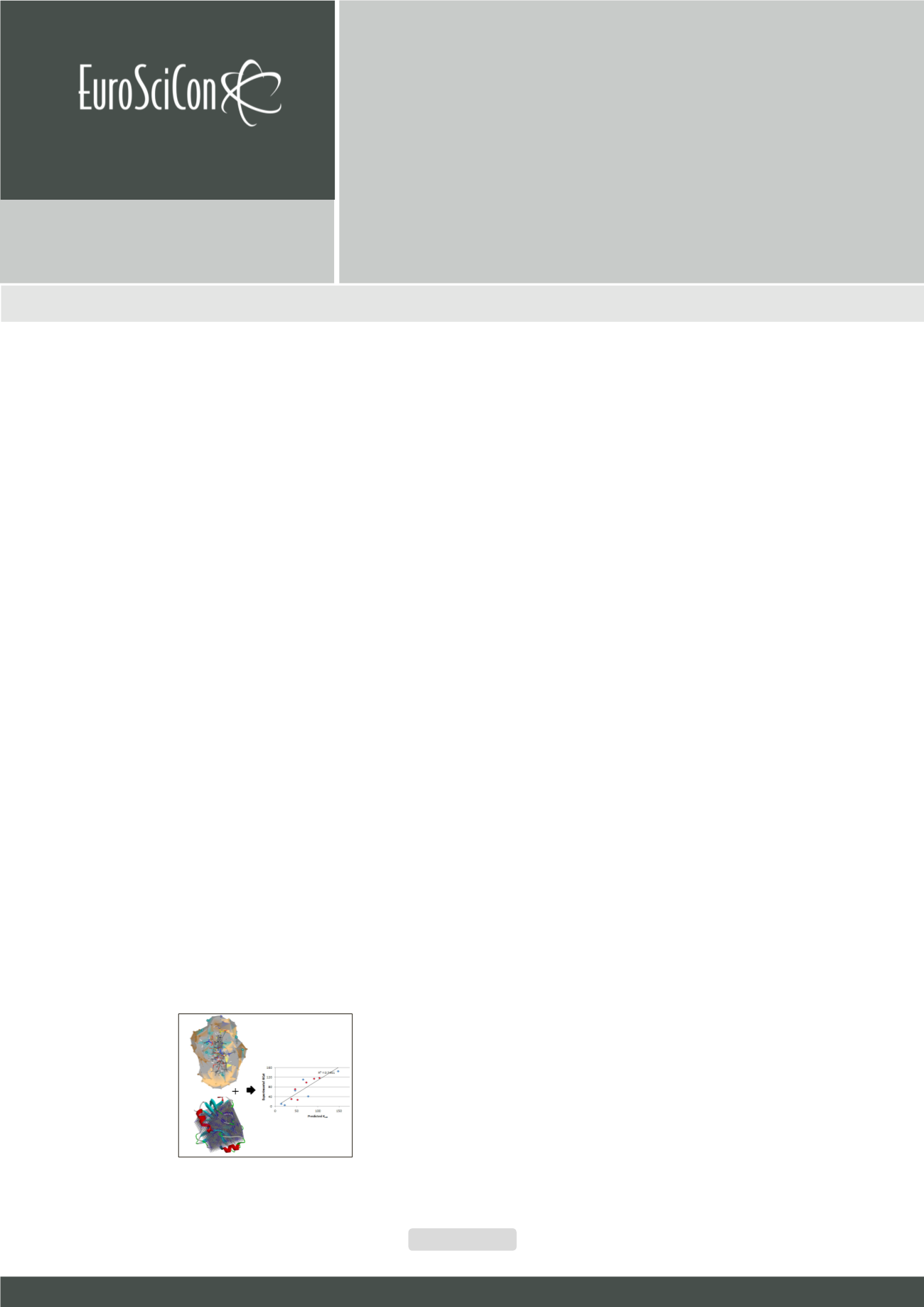
As a proof of concept, for the first time, receptor dependent 4D Quantitative
Structure Activity Relationship (RD-7D-QSAR) to predict kinetic properties of
enzymes has been demonstrated by
Pravin Kumar et al
. The methodology was
extended to study transaminase. Induced-fit scenarios were explored using
QM/MM simulations which were then placed in a grid that stores interactions
energies derived from QM parameters (QM grid). The novelty of this study
is that the mutated enzymes were immersed completely inside the QM grid
and this was combined with solvation models to predict descriptors. After
statistical screening of descriptors, QSAR models showed >90% specificity
and >85% sensitivity towards the experimental activity. Mapping descriptors
on the enzyme structure revealed hotspots important to enhance the
enantioselectivity of the enzyme.
Biography
Pravin Kumar R has completed his Doctorate in Computational
Biology fromBharathiar University, Tamil Nadu, India. He has 15
years of Industrial Experience on different projects pertaining to
target deconvolution and enzyme engineering studies. He has
25 international publications, most of it on techniques such as
Protein Modelling, Molecular Dynamics, Quantum Mechanics
Hybridised with Molecular Dynamics (QM/MM), 4D QSAR,
etc. He has developed the Enzyme Engineering Framework
which is composed of algorithms and screening protocols
of core quantum mechanics, QM/MM and QSAR techniques.
The framework can predict hotspots and enzyme variants with
better activity (K
cat
, K
m
). This framework was used to engineer
transaminase to expand its substrate scope towards bulky
ketones. He has participated and given oral presentation in
Enzyme Engineering conferences: BIOSIG 2014, Toyama,
Japan, BIOSIG 2015 Boston, USA and BIOSIG 2015, Toulouse,
France. He holds several positions such as, Bioinformatician
in VittalMallya Scientific ResearchFoundation, Bangalore,
India Aug (’2004 to Aug’ 2007); Team Head of Research in
Bioinformaticsat Jigsaw Bio Solutions Pvt Ltd., Bangalore,
India (Sep’2007 to Dec’2008); Project head for Computational
Biology at Prescient Biosciences Pvt. Ltd, Peenya, Bangalore,
India (Jan’2009 to Aug’2010); Team lead and Senior Scientist,
in silico, Polyclone Bioservices Pvt Ltd, Jayanagar, Bangalore,
India (Oct’ 2010 to Aug’ 2016) and Director, Quantum Zyme,
Bangalore, India from Sep’ 2016 to May’ 2018. He is the
Reviewer of
Journals J. Biomolecular Structure and Dynamics,
J. Molecular Catalysis, J. Computational Biology and Chemistry
.
pravinpaul2@gmail.com pravin.k@kcat.co.inA novel 7D-QSAR approach, combining QM based grid and
solvation models to predict hotspots and kinetic properties of
mutated enzymes: an enzyme engineering perspective
Pravin Kumar R
1
and Roopa L
2
1
Kcat LLP, Bengaluru, India
2
Mount Carmel College, Bengaluru, India
Pravin Kumar R et al., Insights Enzyme Res 2018, Volume 2
DOI: 10.21767/2573-4466-C1-002
7D-QSAR protocol/paradigm to predict
enzyme kinetic properties
















