
Crystallography 2018
Structural Chemistry & Crystallography Communication
ISSN: 2470-9905
Page 47
June 04-05, 2018
London, UK
3
rd
Edition of International Conference on
Advanced Spectroscopy,
Crystallography and Applications
in Modern Chemistry
T
argeting protein-protein interactions with small molecules
presents a number of well documented challenges, including
the largely flat, featureless interfaces for many published
structures. While structural information has undoubtedly assisted
in drug discovery through suggesting direction and properties
for compound elaboration, the guidance provided can only be
truly valuable if the structure on which it is based is an accurate
representation of the precise conformation of the protein being
targeted. Crystal structures, although visually compelling, may
not represent the biologically relevant conformation of the target,
and can suffer from distortion due, for example, to intermolecular
contacts in the lattice. Orthogonal biophysical techniques, such
as Double Electron-electron Resonance (DEER), in conjunction
with spin labeling at specified points on the surface of target
proteins, can be used to probe natural conformational sampling,
and provide distance measurements, which can be compared to
those obtained from equivalent positions in crystal structures.
Existing crystal structures may thus be adjusted, using advanced
molecular dynamics simulations, to accommodate the distance
measurements from DEER and create working models of target
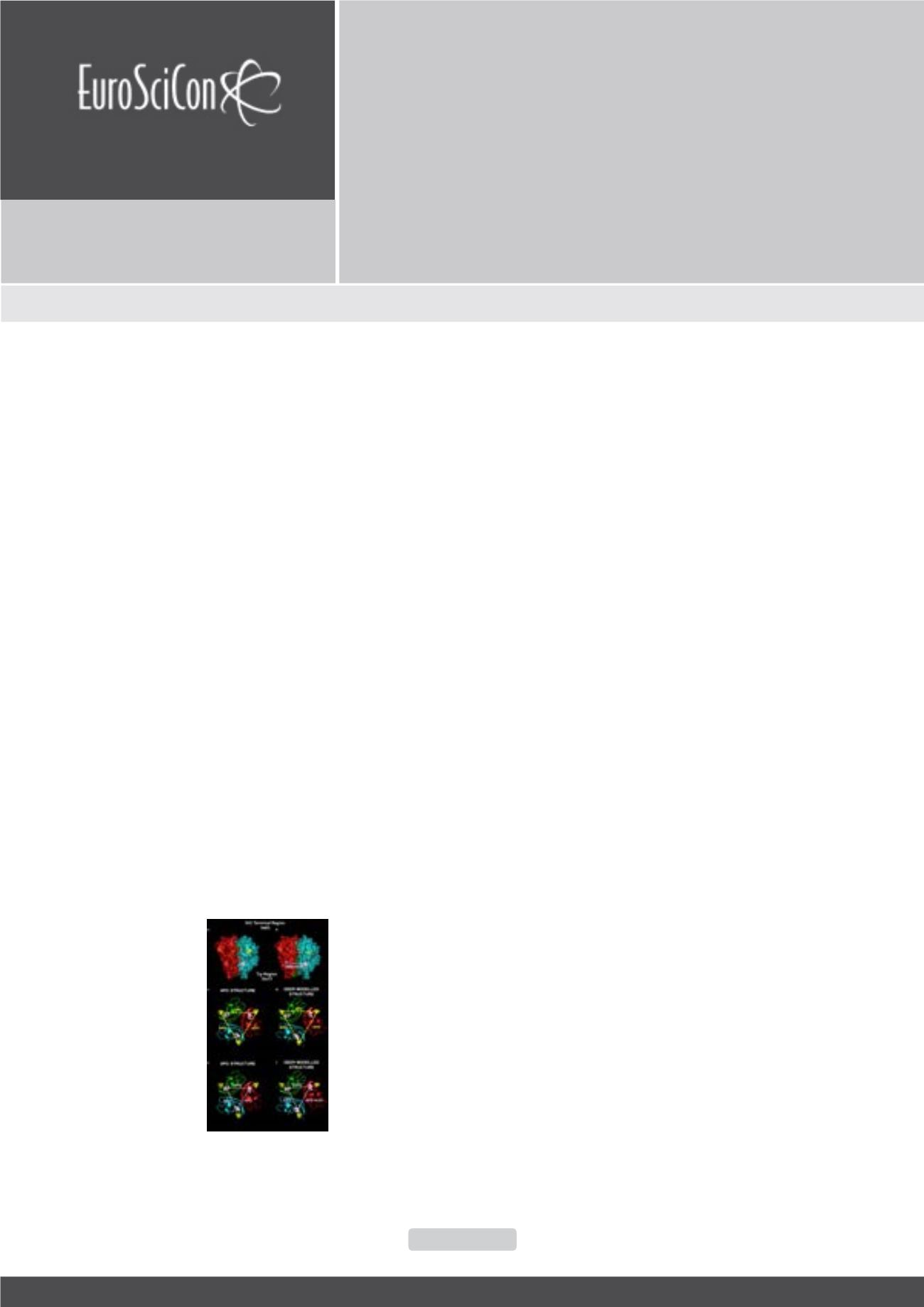
proteins in biologically relevant conformations. For example
the image below shows how a crystal structure of apo TNF was
adjusted using distance data from DEER, to generate a working
model of a new conformation of the target, which may be helpful
in drug discovery.
Recent Publications
1. Importance of Rigidity in Designing Small Molecule
Drugs to tackle Protein-protein Interactions through
stabilization of Desired Conformers. Lawson ADG;
MacCoss M; Heer J. Journal of Medicinal Chemistry
Doi 10.1021/acs.jmedchem.7b01120, 2017
2. Natural Conformational Sampling of Human TNFalpha
Visualized by Double Electron-Electron Resonance.
Carrington B; Myers WK; Horanyi P; Calmiano M; Lawson
ADG. Biophysical Journal. 113(2):371-380, 2017
3. Combining Molecular Scaffolds from FDA Approved
Drugs: Application to Drug Discovery. Taylor RD;
MacCoss M; Lawson AD. Journal of Medicinal
Chemistry. 60(5):1638-1647, 2017
4. Computational design of an epitope-specific Keap1
binding antibody using hotspot residues grafting and
CDR loop swapping. Liu X; Taylor RD; Griffin L; Coker SF;
Adams R; Ceska T; Shi J; Lawson AD; Baker T. Scientific
Reports. 7:41306, 2017.
5. Small Molecule Targeting of Protein-Protein Interactions
through Allosteric Modulation of Dynamics. Cossins
BP; Lawson AD. Molecules. 20(9):16435-45, 2015
6. Rings in drugs. Taylor RD. MacCoss M. Lawson AD.
Journal of Medicinal Chemistry. 57(14):5845-59, 2014
7. Antibody-enabled small-molecule drug discovery.
Lawson AD. Nature Reviews Drug Discovery. 11(7):519-
25, 2012.
Biography
Alastair has been closely involved with the discovery of UCB/Celltech’s thera-
peutic antibodies, including Mylotarg®, Besponsa®, Cimzia®, romosozumab,
dapirolizumab pegol, olokizumab, bimekizumab and UCB7665. Alastair led the
development of UCB’s proprietary antibody variable region discovery platform,
and is now applying structure-based, rational design to antibody discovery. He
pioneered UCB’s small molecule protein/protein interaction initiative, in which in-
formation derived from antibodies is applied to the discovery and design of new
chemical entities. Current research interests include the use of function-modify-
ing antibody fragments to define specific conformations of target proteins, link-
ing X-ray crystallography, orthogonal biophysical techniques, molecular dynam-
ics simulations and antibody technology to small molecule fragment screening
alastair.lawson@ucb.comANTIBODY-ENABLED SMALL MOLECULE DRUG DISCOVERY
Alastair Lawson
UCB, Slough, UK
Alastair Lawson, Struct Chem Crystallogr Commun 2018, Volume 4
DOI: 10.21767/2470-9905-C1-005
















