Zenobia: CODIS 13 STR Loci Allele Detection Tool
Osamah S Alrouwab1,2*, Esraa B. Algblawi2, Moudah B. Kareem2, Sabreen A. Aboujildah2, Magdi A. Allafi2 and Mahmoud Gargotti 3
1Department of Biochemistry, Faculty of Medicine, Sbratha University, Sbratha, Libya
2Department of Biotechnology, Aljafra University, Alsahla, Libya
3Department of Microbiology, Faculty of Medicine, University of Zawia, Zawia, Libya
- *Corresponding Author:
- Osamah S Alrouwab
Department of Biochemistry,
Faculty of Medicine, Sbratha University, Sbratha,
Libya,
Tel No: 218913106276;
E-mail: usamaerawab@gmail.com
Received: February 18, 2022, Manuscript No. IPGMBR-22-11455; Editor assigned: February 21, 2022, PreQC No. IPGMBR-22-11455 (PQ); Reviewed: March 07, 2022, QC No. IPGMBR-22-11455; Revised: March 11, 2022, Manuscript No. IPGMBR-22-11455 (R); Published: March 18, 2022, DOI: 10.36648/ipgmbr.22.6.68
Citation: Alrouwab OS, Algblawi EB, Kareem MB, Aboujildah SA, Allafi MA et al. (2022) Zenobia: CODIS 13 STR Loci Allele Detection Tool. Genet Mol Biol Vol:6 No:1
Abstract
Short Tandem Repeats (STRs) are one of the utmost mutable provinces in the human genome. They comprise tandem repeating DNA sequences ranging in length from two to six base pairs. Owing to their significant mutation rate, they exhibit considerable variation in pattern among populations and the capacity to be passed on from generation to generation. These loci are broadly employed in medicine, biology, and criminal investigation. They are pivotal in the genesis of a variety of genetic illnesses and have been intensively investigated in forensics, population genetics, and genetic genealogy. Although many implementations that manage STR loci are offered, the overwhelming majority of them rely primarily on the Command-Line Interface (CLI) inputs, which frequently necessitate the implementation of tools carried out in various scripting languages. Installing and launching programs through the Command Line (CL) is timeconsuming and/or unprofitable for many students and scholars. The fundamental intention of this project is to develop a cross-platform Graphical User Interface (GUI) package directed to the Combined DNA Index System (CODIS) STR analysis. Zenobia is a Java-based application considered as a step in consistently making CL-only programs available to more apprentices and researchers. In general, Zenobia's application outcomes satisfy the evaluation metrics for efficiency and time consumption. However, more genetic markers should be introduced to increase productivity of the application.
Keywords
Short tandem repeats; Java; Combined DNA index system; Command line; Forensics
Introduction
Revolutionary, Genetic fingerprinting is one of the emerging technologies that has drastically influenced the realm of forensic medicine and has profoundly altered forensic evidence forever [1]. DNA fingerprinting (DNA profiling or forensic genetics are synonyms also used to designate the same methodology) provides a comparative analysis of DNA to solve legal problems that include paternity tests, the identification of individuality in criminal proceedings in which biological evidence is discovered at crime scenes, and distinguishing the victims of major disasters from the remains [2-3]. Historically, in the mid-eighties of the past century, a research team from the University of Leicester, UK, led by the founder of DNA fingerprinting, Sir Alec Jeffrey's, established the era of using DNA in forensic evidence [4]. The microsatellite or Short Tandem Repeats (STRs) markers have been the most extensively used approach for detecting DNA profiles [5]. They are ubiquitous throughout the DNA and reside on average 6-10 kb apart [6-7]. Attributed to their density, polymorphism, and PCR amplification, STRs were measured as reliable biomarkers for genomic mapping and genetic linkage assessment [8-9]. DNA profiling based on STR PCR amplification has the benefit of being more responsive than traditional methods. In addition, their negligible allele size (typically<300 bp) makes the STR system more likely to succeed with older or poorly preserved samples containing only degraded DNA [10-12]. It has been over four decades since the FBI Laboratory selected thirteen STR genetic markers for what is now known as the Combined DNA Index System (CODIS) [13-15]. The CODIS loci used in the US are TPOX, VWA, D3S1358, CSF1PO, FGA, TH01, D13S317, D16S539, D18S51, D5S818, D7S820, D8S1179, and D21S11 [16]. These loci have become the conventional coinage of information exchange for verifying human identity for both judicial case studies and paternity testing due to their accessibility and utilization in the form of commercial STR kits [17-18]. Addressing profile sequence data is a struggle for many students and researchers [19]. Despite, a wide range of programs capable of analyzing STR loci being available, all of them rely on the Command-Line Interface (CLI) commands or are not specifically directed at DNA markers used in forensic investigations. Moreover, they often rely on a set of complementary tools that are implemented in various script languages [20-23]. Some legacy applications for finding tandem repeats within a sequence include: Mreps, demonstrated by Kolpakov Roman and Gregory Kucherov (2003), it’s a sophisticated software for detecting tandem repeated structures in DNA sequences. Mreps could indeed detect all sorts of tandem repeats in a single run on an entire genomic sequence. It has a resolution setting that enables the software to detect 'fuzzy' repetitions [24]. Marco Pellegrini and Alessio Vecchio (2010) developed TRStalker, an algorithm (christened TRStalker) with the intent of discovering Tandem Repeats (TRs) that are hard to identify, owing to their characteristic fuzziness, which is attributed to the high rates of base substitutions, insertions, and deletions [25]. In 2010, Pokrzywa, Rafal, and Andrzej Polanski introduced the Burrows–Wheeler Tandem Repeat Searcher (BWTRS). It is an online web-based utility that scans for specific instances of tandem repeats in DNA sequences, BWTRS adopts the block-sorting compression algorithm [26]. In this paper, we intend to provide a novel tool capable of detecting and determining the numbers of alleles of CODIS loci stored in a plain text FASTA format.
Materials and methods
Zenobia v1.0
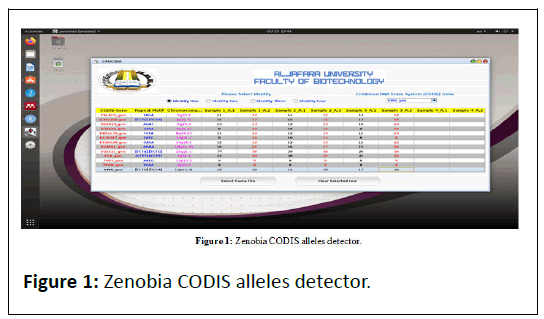
Zenobia (Figure 1) is a Java-based Graphical User Interface (GUI) tool, for CODIS 13 Alleles detection released under the GNU General Public License. The source code is freely available on GitHub.
Dataset
Zenobia core dataset imported from STR base, a public dataset provided by the National Institute of Standards and Technology during September 2021. Only CODIS 13 STR markers data were chosen, namely, CSF1PO, FGA, TH01, TPOX, vWA, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, and D21S11 (Table 1).
| Locus | Repeat motif | Repeat category | Chromosome location | Allele range |
|---|---|---|---|---|
| CSF1PO | AGAT | Simple | 5q33.1 | 16-May |
| FGA | [CTTT] [CCTT] | Compound | 4q31.3 | 12.2-51.2 |
| TH01 | TCAT | Simple | 11p15.5 | 14-Mar |
| TPOX | TGAA | Simple | 2p25.3 | 16-Apr |
| VWA | [TCTG] [TCTA] | Compound | 12p13.31 | 25-Oct |
| D3S1358 | [TCTG] [TCTA] | Compound | 3p21.31 | 20-Aug |
| D5S818 | AGAT | Simple | 5q23.2 | 18-Jun |
| D7S820 | GATA | Simple | 7q21.11 | 16-May |
| D8S1179 | [TCTA] [TCTG] | Compound | 8q24.13 | 20-Jul |
| D13S317 | TATC | Simple | 13q31.1 | 17-May |
| D16S539 | GATA | Simple | 16q24.1 | 16-Apr |
| D18S51 | AGAA | Simple | 18q21.33 | 7-39.2 |
| D21S11 | [TCTA] [TCTG] | Complex | 21q21.1 | 12-41.2 |
Table 1: Common STR loci.
Case scenario and input files
A paternity dispute case based on matches of the alleles at the CODIS 13 STR loci between a child and mother and alleged father (trio cases), from the Arab Republic of Egypt in 2012 (Table 2), documented by Mr. Sherif H. El-Alfy, used to simulate and construct dummy profile files [27].
| STR locus | Child | Mother | Alleged father |
|---|---|---|---|
| D3S1358 | 15,17 | 15,16 | 17,18 |
| D5S818 | 13,13 | 12, 13 | 10, 13 |
| D7S820 | 8,10 | 10,10 | 8,10 |
| D8S1179 | 11,12 | 12,13 | 11,13 |
| D13S317 | 8, 10 | 10, 13 | 8,8 |
| D16S539 | 12, 12 | 12, 13 | 11,12 |
| D18S51 | 16, 16 | 16,17 | 15,16 |
| D21S11 | 30,30 | 29,30 | 29,30 |
| FGA | 23,24 | 20,24 | 21,23 |
| TH01 | 9,9 | 8,9 | 8,9 |
| TPOX | 8,8 | 8,8 | 8,8 |
| VWA | 18,20 | 14,18 | 17,20 |
| CSF1PO | 11, 12 | 11,12 | 12, 12 |
Table 2: Typing results of 13 autosomal STR loci analysis.
Since the program support only a plain text Fasta file format. To evaluate the performance of the tool we generate dummy files that contain random sequences with real allelic varia ion sequences imported from the Entrez database provided by The National Center for Biotechnology Information (NCBI) for locusspecific information (Table 3).
| Child | Mother | Alleged father | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Allele one | Allele two | Allele one | Allele two | Allele one | Allele two | ||||||
| Allele no. | Accession | Allele no. | Accession | Allele no. | Accession | Allele no. | Accession | Allele no. | Accession | Allele no. | Accession | |
| D3S1358 | 15 | MW218622 | 17 | MK990349 | 15 | MW218622 | 16 | MH166976 | 17 | MK990349 | 18 | MK990350 |
| D5S818 | 13 | MZ325899 | 13 | MZ325899 | 12 | MH167008 | 13 | MZ325899 | 10 | MH166998 | 13 | MZ325899 |
| D7S820 | 8 | MH167026 | 10 | MZ325981 | 10 | MZ325981 | 10 | MZ325981 | 8 | MH167026 | 10 | MZ325981 |
| D8S1179 | 11 | MH105190 | 12 | MH105195 | 12 | MH105195 | 13 | MH105201 | 11 | MH105190 | 13 | MH105201 |
| D13S317 | 8 | MZ325902 | 10 | MK295189 | 10 | MK295189 | 13 | MT298696 | 8 | MZ325902 | 8 | MZ325902 |
| D16S539 | 12 | MT298697 | 12 | MT298697 | 12 | MT298697 | 13 | MW218608 | 11 | MH167254 | 12 | MT298697 |
| D18S51 | 16 | MW218634 | 16 | MW218634 | 16 | MW218634 | 17 | MK569958 | 15 | MW218632 | 16 | MW218634 |
| D21S11 | 30 | MZ325991 | 30 | MZ325991 | 29 | MZ325935 | 30 | MZ325991 | 29 | MZ325935 | 30 | MZ325991 |
| FGA | 23 | MZ325919 | 24 | MH232622 | 20 | MH232609 | 24 | MH232622 | 21 | MH232611 | 23 | MZ325919 |
| TH01 | 9 | MH085123 | 9 | MH085123 | 8 | MW218611 | 9 | MH085123 | 8 | MW218611 | 9 | MH085123 |
| TPOX | 8 | MG988075 | 8 | MG988075 | 8 | MG988075 | 8 | MG988075 | 8 | MG988075 | 8 | MG988075 |
| VWA | 18 | MW218658 | 20 | MH167102 | 14 | MH167077 | 18 | MW218658 | 17 | MW218657 | 20 | MH167102 |
| CSF1PO | 11 | MH085186 | 12 | MN983119 | 11 | MH085186 | 12 | MN983119 | 12 | MN983119 | 12 | MN983119 |
Table 3: Allele number and accession used to evaluate the performance.
Implementation
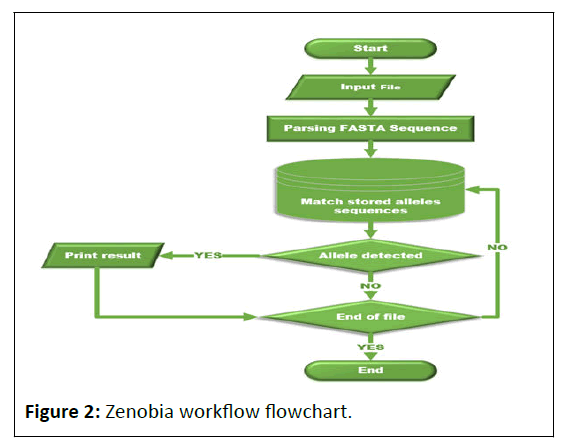
The benchmarks were carried out on personal computers with intel core i5-3470, 3.20 GHz, 16.00 GB of RAM, Linux Ubuntu-20.04.3 64 bit. Zenobia was written in Java programing language using Oracle Java SE Development Kit 11 and Apache NetBeans IDE 12.1 Detection of the allelic type for each STR gene. Zenobia recruits the so-called brute force algorithm to match stored allele patterns to detect locus names and allele numbers (Figure 2).
Results
A total of 78 alleles participated in the experiment, 61.5% of whom are representatives of a simple STR subgroup. Furthermore, 30.1% and 7.7% of candidates engaged with compound and complex STR subgroup correspondingly. The child profile's allele numbers fluctuated from 11 to 30, the mother profile's allele ranged from 8 to 29, while the alleged father allele numbers spanned from 8 to 30. The observed genotype for child profile was, D3S1358 (15,17), D5S818 (13,13), D7S820 (8,10), D8S1179 (11,12), D13S317 (8,10), D16S539 (12,12), D18S51 (16,16), D21S11 (30,30), FGA (23,24), TH01 (9,9), TPOX (8,8), VWA (18,20), CSF1PO (11,12). While mother shows, D3S1358 (15,16), D5S818 (12,13), D7S820 (10,10), D8S1179 (12,13), D13S317 (10,13), D16S539 (12,13), D18S51 (16,17), D21S11 (29,30), FGA (20,24), TH01 (8,9), TPOX (8,8), VWA (14,18), CSF1PO (11,12). Finally, the alleged father records, D3S1358 (17,18), D5S818 (10,13), D7S820 (8,10), D8S1179 (11,13), D13S317 (8,8), D16S539 (11,12), D18S51 (15,16), D21S11 (29,30), FGA (21,23), TH01 (8,9), TPOX (8,8), VWA (17,20), CSF1PO (12,12).
Discussion
The purpose of this study was to develop a multi-platform, user-friendly, and open-source CODIS 13 STRs allele detector. Many methods for locating short tandem repeats over DNA sequences have been developed in response to their relevance in understanding STR loci [28]. Some tools are out of date, and a handful of them are no longer accessible [29]. There are, however, several programs available that operate either on the command line or as standalone web services. In this section, different tools will be surveyed for their capabilities to detect STR loci. TAREAN, a command-line, computational approach for automatically detecting satellite repeats in unassembled Next- Generation Sequencing (NGS) sequences. TAREAN is built with customized Python and R packages, to discover new satellite repeats, which were then confirmed on metaphase chromosomes using FISH with probes generated based on reconstructed monomer sequences [30]. STRetch, a commandline tool written as python scripts directed to the analysis of STRs from Whole-Genome-Sequencing (WGS) results, was developed by Harriet, et al. (2018). TRetch seems to have a low False Discovery Rate (FDR) for deleterious STR expansions related to Mendelian disorder, It is designed for STR linked to genetic disorders [31]. TandemTools, a python-based tool developed by Mikheenko, Alla, et al. (2020) detected Extra-Long Tandem Repeats (ETRs) [32]. Contrarily, in comparison to other comparable programs, Zenobia adopts an entirely different approach. None of the tandem repeats detecting algorithms were implemented since the program's objective is to determine the allele number associated with each locus, not only the existence or absence of these repeats. This grants Zenobia an edge over other current programs, which are only capable of spotting tandem repetitions.
Genotyping criteria in Zenobia
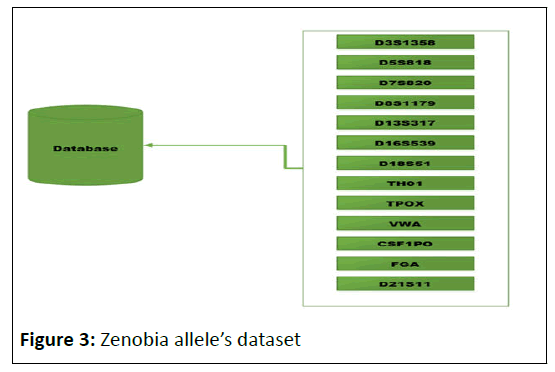
Zenobia was implemented to identify readings for pre-defined CODIS 13 STR loci. For this aim, 13 distinct classes representing the major positions loci have been constructed, and each of them maintains the dataset of its alleles as described by the National Institute of Standards and Technology (Figure 3).
The brute force algorithm was used to achieve a perfect match between the alleles stored in the database, validate their appearance, and identify the precise number of the corresponding allele. It is regarded as one of the most logical choices for the string pattern-matching challenge. Simply matching the pattern in the target at consecutive positions from left to right is the focus of this method. If the comparison window fails, it shifts one letter to the right until the end of the target sequence is attained. Despite the algorithm's poor theoretical performance, our measurements show that it is one of the fastest techniques when the pattern is a short sequence.
Limitation of Zenobia
Zenobia supports only one type of file format, the so-called FASTA. Furthermore, the stored datasets do not only contain complementary sequences of the alleles.
Conclusions
We designed a Bioinformatics application using JAVA language version 11. It enables us in interpreting FASTA files, identify CODIS 13 loci, and determine the allelic number from a nucleotide sequence. Zenobia has done an excellent job at applying the boundary values in terms of precision and time consumption. When reading the 78 allelic profiles, no faults were encountered. However, additional STR loci are still required to be added.
References
- J Hilbert (2018) The disappointing history of science in the courtroom: Frye Daubert and the ongoing crisis of junk science in criminal trials Okla. L Rev 71:759
- KRD Nurse (2018) Forensic Experts’ Best Practices in DNA Collection Analysis and Testimony: A Delphi Study
- A Gang, VK Shrivastav (2020) Single-Nucleotide Polymorphism: A Forensic Perspective Handb. DNA Profiling 1–22
- J A Bright, H Kelly, Z Kerr, C McGovern, D Taylor et al. (2020) The interpretation of forensic DNA profiles: an historical perspective. JR Soc New Zeal 50:211–225
- Tao R, Wang S, Zhang J, Zhang J, Yang Z, et al. (2018) Separation/extraction, detection, and interpretation of DNA mixtures in forensic science. Int J legal med132(5):1247-1261.
[Crossref] [Google Scholor] [Pubmed]
- C Francastel, F Magdinier (2019) DNA methylation in satellite repeats disorders Essays. Biochem 63:757–771
[Crossref] [Google Scholor] [Pubmed]
- G Xu, J Lyu, Q Li, H Liu, D Wang, et al. (2020) Evolutionary and functional genomics of DNA methylation in maize domestication and improvement. Nat Commun 11:1-12
[Crossref] [Google Scholor] [Pubmed]
- Weymaere J, Vander Plaetsen AS, Tilleman L, Tytgat O, Rubben K, et al. (2020) Kinship analysis on single cells after whole genome amplification. Sci rep 10(1):1-9.
[Crossref] [Google Scholor] [Pubmed]
- WS Al-Qahtani, TM Al-Hazani, FA Safhi, MA Alotaibi, DM Domiaty et al. (2021) Assessment of Metastatic Colorectal Cancer (CRC) Tissues for Interpreting Genetic Data in Forensic Science by Applying 16 STR Loci among Saudi Patients Asian Pacific. J Cancer Prev 22 2797–2806
[Crossref] [Google Scholor] [Pubmed]
- Lynch c, Fleming R (2019) A review of direct polymerase chain reaction of DNA and RNA for forensic purposes Wiley Interdiscip. Rev Forensic Sci 1:1335
- Nigam K, Srivastava A, Sahoo S, Dubey IP, Tripathi IP, et al. (2020) Sequential Advancements of DNA Profiling: An Overview of Complete Arena Forensic DNA Typing. Princ Appl Adv 45–68
- Thakar M, Joshi B, Shrivastava P (2020) Usefulness of Mini-STRs in Analyzing Degraded DNA Samples and Their Forensic Relevance in: Forensic DNA Typing. Princ Appl Adv Springer 205–222
- Katsanis SH (2020) Pedigrees and perpetrators: Uses of DNA and genealogy in forensic investigations. Annu Rev Genomics Hum Genet 21:535–564
[Crossref] [Google Scholor] [Pubmed]
- Kitnick J (2019) Killer’s Code: Familial DNA Searches Through Third-Party Databases under Carpenter Cardozo. L Rev 41:855
- Neuvonen A (2017) Finnish population genetics in a forensic context
- Huang X (2019) Short Tandem Repeat Profiles In Ovarian Carcinoma Cells During Primary Culture
- Kaushik S, Sahajpal V (2020) Capillary Electrophoresis Issues in Forensic DNA Typing, in: Forensic DNA Typing. Princ Appl Adv Springer 223–238
- NF de Groot, BC van Beers, MeynenG (2021) Commercial DNA tests and police investigations: a broad bioethical perspective. J Med Ethics
[Crossref] [Google Scholor] [Pubmed]
- Meng T, Soliman AT, Shyu ML, Yang Y, Chen SC, et al. (2013) Wavelet analysis in current cancer genome research: a survey, IEEE/ACM Trans. Comput Biol Bioinforma 10:1442–14359
[Crossref] [Google Scholor] [Pubmed]
- Christopher FE, Myers KJ (2018) Siem-Enabled Cyber Event Correlation (What And How). NAVAL POSTGRADUATE SCHOOL MONTEREY CA
- L Codó Tarraubella (2019) Computational Infrastructures for biomolecular research
- Boattini A, Sarno S, Mazzarisi AM, Viroli C, S De Fanti, et al. (2019) Estimating Y-str Mutation Rates and tmrca through Deep-Rooting Italian pedigrees. Sci Rep 9:1–12
[Crossref] [Google Scholor] [Pubmed]
- Gettings KB, Borsuk LA, Steffen CR, Kiesler KM, Vallone PM (2018) Sequence-based US population data for 27 autosomal STR loci Forensic. Sci Int Genet 37:106–115
[Crossref] [Google Scholor] [Pubmed]
- Kolpakov R, Bana G, Kucherov G (2003) Mreps: Efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res 31:3672–3678
[Crossref] [Google Scholor] [Pubmed]
- Pellegrini M, Renda ME, Vecchio A (2012) Tandem repeats discovery service (TReaDS) applied to finding novel cis-acting factors in repeat expansion diseases. BMC Bioinformatics 13:1–15
[Crossref] [Google Scholor] [Pubmed]
- Pokrzywa R, Polanski A (2010) BWtrs: a tool for searching for tandem repeats in DNA sequences based on the Burrows–Wheeler transform. Genomics 96:316–321
[Crossref] [Google Scholor] [Pubmed]
- S El-Alfy, A El-Hafez (2012) Paternity testing and forensic DNA typing by multiplex STR analysis using ABI PRISM 310 Genetic Analyzer. J Genet Eng Biotechnol 10:101–112
- Halman A (2021) Advancing the detection of short tandem repeats in health and disease
- Parisi V, V De Fonzo, Aluffi-Pentini F (2003) STRING: finding tandem repeats in DNA sequences. Bioinformatics 19:1733–1738
[Crossref] [Google Scholor] [Pubmed]
- Novák P, Ávila Robledillo L, Koblížková A, Vrbová I, Neumann P, et al. (2017) TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res 45:111–111
[Crossref] [Google Scholor] [Pubmed]
- Dashnow H, Lek M, Phipson B, Halman A, Sadedin S, et al. (2018) STRetch: detecting and discovering pathogenic short tandem repeat expansions. Genome Biol 19:1–13
[Crossref] [Google Scholor] [Pubmed]
- A Mikheenko A, Bzikadze AV, Gurevich A, Miga KH, Pevzner PA, et al. (2020) TandemTools: mapping long reads and assessing/improving assembly quality in extra-long tandem repeats. Bioinformatics 36:75–83
[Crossref] [Google Scholor] [Pubmed]
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences