The Occurrence of Multidrug Resistant E. Coli which Produce ESBL and Cause Urinary Tract Infections
Talat I Elsayed1, Hala AF Ismail2, Samir A Elgamal2 and Ahmed HA Gad1*
1Botany Department, Benha University, Qalubia, Egypt
2Medical Microbiology & Immunology Department, Tanta University, Tanta, Egypt
- *Corresponding Author:
- Ahmed HA Gad
Faulty of Science, Botany Department
Microbiology Branch, Benha University
Benha, Qalubia, Egypt
Tel: 033911598
E-mail: abohamdy009@gmail.com
Received Date: April 26, 2017; Accepted Date: May 09, 2017; Published Date: May 16, 2017
Citation: Elsayed TI, Ismail HA, Elgamal SA, et al. (2017) The Occurrence of Multidrug Resistant E. Coli which Produce ESBL and Cause Urinary Tract Infections. J Appl Microbiol Biochem. Vol. 1 No. 2:8
Abstract
Urinary tract infection (UTI) is caused by Gram-negative bacteria such as Escherichia coli, Klebsiella species, Enterobacter species, Proteus species and Gram-positive bacteria like Enterococcus species, and Staphylococcus saprophyticus. E. coli is the most common organism causing both community as well as hospital acquired UTI.
The aim of this study was to determine the occurrence of multidrug resistant strains among UTI caused by E. coli isolates against commonly used antimicrobial agents and the possible role of extended spectrum beta-lactamases (ESBL) in E. coli resistance to antibiotics.
One hundred E. coli isolates from 262 inpatients were identified. Susceptibility to various antibiotics was checked using standard methods. All Isolates were examined for the presence of ESBL using combination disc method, indicating ESBL production.
Most of isolates were resistant to ampicillin and cephalothin. Overall resistance to beta-lactam antibiotics ampicillin and cephalothin was 95% and 93% respectively. Almost all Isolates were sensitive to imipenem with 2% resistance. Thirty six per cent of E. coli isolates were ESBL producers.
In order to find out the plasmid profile of these isolates, plasmid preparations were made for each isolate. Out of 100 clinical isolates 68 of them were found to have one or more plasmids responsible for ESBL resistance.
Our results conclude that most of E. coli isolated during this study, are ESBL producing. It is highly recommended that antibiotic prescription should be monitored according to the guidelines. Antibiotic consumption should be monitored both in healthcare facilities as well as in community. The role of Infection prevention and control is crucial in all healthcare facilities to decrease the occurrence of antibiotic resistance.
Keywords
Urinary tract infection; Multidrug resistant E. coli; ESBL; Plasmid profile
Introduction
Extended-spectrum β-lactamase-producing Enterobacteriaceae is an emerging public health concern. Complications in UTIs have increased because of the prevalence of extended spectrum beta-lactamases (ESBL) producing bacterial pathogens which are also causing many management and epidemiological issues [1]. These complications came out from the increasing resistance among UTI pathogens to conventional drugs. The most common etiological agent in UTI is E. coli and the prevalence of ESBLs producing E. coli isolated from community acquired urinary tract infection came out to be 17% and for urinary tract nosocomial infection was 58% [2].
ESBL production confers resistance to all the beta-lactam antibiotics, except carbapenems and cephamycins. In addition, ESBL encoding plasmids also carry genes which encode resistance to other class of antibiotics such as fluoroquinolones, aminoglycosides and sulfonamides [3]. It was proved that resistance to penicillin, sulfamethoxazole, trimethoprime and cephalothin were 100%, 30.89%, 16.26% and 20.32%, respectively [4]. Attempts have been made to decrease the prevalence of ESBL producing organisms by substituting earlier cephalosporins with a fourth-generation cephalosporin or beta-lactam/betalactamase inhibitor combinations [5]. Therefore, there is a need, to determine the current prevalence and phenotype of multidrug resistant strains among UTI caused by E. coli isolates against commonly used antimicrobial agents [6].
Much of the problem of antimicrobial resistance has been shown to be due to the presence of transferable plasmids encoding multidrug resistance and their dissemination among different enterobacterial species [7] and it is common for a single plasmid to simultaneously mediate resistance to multiple antimicrobials and to be shared among different bacterial genera. In plasmid profiling, isolates usually vary in size and number. For [8] out of the 99 E. coli pathotypes, 35 (35.4%) were found to possess plasmids, which ranged in sizes from 1.7 kb to 4.5 kb.
The present study was conducted to achieve resistance profile among E. coli clinical isolates from our local area in Tanta hospitals against commonly prescribed antibiotics. Further analysis was done to identify the prevalence of multidrug resistant strains and the possible role of ESBL in E. coli resistance to antibiotics. Plasmid profiling was done to know the role and nature of plasmid as an antibiotic resistance marker in ESBLs.
Materials and Methods
Isolation and identification of E. coli
Bacterial strains: A total of 100 isolates of E. coli from urinary tract infected patients were studied. The isolates were nonrepetitive and were obtained consecutively from urine specimens from inpatients in urology wards of Tanta university hospitals in the period between June 2013 and September 2014.
Isolation of pathogens: Pathogens were isolated using standard media, including Cystine lactose electrolyte deficient (CLED) agar, blood agar and MacConkey‘s agar. Specimens were inoculated using standard techniques. Plates were incubated overnight at 37°C before the plates were inspected for growth [9].
Identification of isolates: Isolates were identified based on morphological and biochemical characteristics. Morphological characters include colony diameter, shape, texture, height, edge, color and reaction on standard media including CLED agar, Eosin methylene blue (EMB) agar and MacConkey‘s agar. Gram staining for all isolates was done. Routine biochemical tests i.e., lactose fermentation, indole production, methyl red, Voges-Proskauer, citrate utilization, urease and motility were tested according to Berg’s manual [10].
Antimicrobial susceptibility
Antimicrobial susceptibility was determined by using Kirby-Bauer disk diffusion technique, recommended by clinical and Laboratory Standards Institute CLSI, guidelines. Antimicrobial agents used in this study were: Ampicillin (AMP), Sulphamethoxazole/ Trimethoprime (SXT), Nalidixic acid (NA), Norfloxacin (NOR), Gentamicin (CN), Nitrofurantoin (F), Imipenem (IPM), Cephalothin (KF). Discs were purchased from Oxoid. Susceptibility test was done using method of [11].
Detection of ESBL producers by combination disc method
Testing for ESBL production was carried out by inoculating bacterial suspension of the isolate with a turbidity equivalent to 0.5 McFarland Standard on Muller-Hinton agar in accordance with NCCLS M100-S10 (M2) guidelines for disc diffusion testing. Separate commercial discs containing cefotaxime (CTX 30 μg) and ceftazidime (CAZ 30 μg) alone and with clavulanic acid (10 μg) were used. Antibiotic discs were placed over the lawn culture according to the test performed.
Two tests were done; initial screen test and phenotypic confirmatory test. According to Clinical & Laboratory Standards Institute CLSI; a zone of ≤ 27 mm for cefotaxime and ≤ 22 mm for ceftazidime indicate ESBL production as a positive.
ESBL initial screen test: An increase in zone size of ≥ 5 mm for cefotaxime and ceftazidime alone and with clavulanic acid was considered to confirm ESBL producing strain as a positive ESBL phenotypic confirmatory test.
Plasmid profiling
Plasmid profile of all clinical isolates was determined. Isolation of plasmids was carried out by alkaline lysis method [12]. DNA gel electrophoresis was done using 0.8% agarose gel [13]. Pure Yield™ Plasmid Miniprep System was used for DNA purification by centrifugation through three main steps; preparing lysate, washing and elution. A 600 μl of bacterial culture was added to 1.5 ml microcentrifuge tube and then 100 μl of Cell Lysis Buffer (Blue) and centrifuged at maximum speed. Flow through was discarded after transfering the supernatant to a PureYield™ Minicolumn. Washing was done with 200 μl of Endotoxin Removal Wash then 400 μl of Column Wash Solution was added and centrifuged. Finally; elution was done with 30 μl of Elution Buffer. Gel electrophoresis was done through three main steps; preparation, loading and running the gel and post-electrophoresis staining.
Results
Identification of isolates
All isolates were identified as E. coli based on their morphological and biochemical characters.
Antimicrobial susceptibility
Isolates were all tested against 8 antibiotics included in the study. Table 1 indicates the resistance pattern of clinical isolates against 8 antimicrobial agents. The overall antimicrobial resistance patterns of the isolates against different antibiotics were as follows: Ampicillin 95%, Sulphamethoxazole/Trimethoprime 69%, Nalidixic acid 70%, Norfloxacin 59%, Gentamicin 31%, Nitrofurantoin 16%, Cephalothin 93% and Imipenem 2%.
Table 1 Resistance pattern of clinical isolates from UTI (n=100) against antibiotics.
| Antibiotic | Clinical isolates from | ||
|---|---|---|---|
| resistance | urinary tract infection | ||
| pattern | (n=100) | ||
| n | % | ||
| ▪AMP 10 ug | |||
| Resistant | 95 | 95.0 | |
| Sensitive | 3 | 3.0 | |
| Intermediate | 3 | 3.0 | |
| ▪SXT 25 ug | |||
| Resistant | 69 | 69.0 | |
| Sensitive | 28 | 28.0 | |
| Intermediate | 3 | 3.0 | |
| ▪NA 30 ug | |||
| Resistant | 70 | 70.0 | |
| Sensitive | 22 | 22.0 | |
| Intermediate | 8 | 8.0 | |
| ▪NOR 10 ug | |||
| Resistant | 59 | 59.0 | |
| Sensitive | 37 | 37.0 | |
| Intermediate | 4 | 4.0 | |
| ▪CN 10 ug | |||
| Resistant | 31 | 31.0 | |
| Sensitive | 59 | 59.0 | |
| Intermediate | 10 | 10.0 | |
| ▪F 300 ug | |||
| Resistant | 16 | 16.0 | |
| Sensitive | 65 | 65.0 | |
| Intermediate | 19 | 19.0 | |
| ▪KF 30 ug | |||
| Resistant | 93 | 93.0 | |
| Sensitive | 4 | 4.0 | |
| Intermediate | 3 | 3.0 | |
| ▪IPM 10 ug | |||
| Resistant | 2 | 2.0 | |
| Sensitive | 96 | 96.0 | |
| Intermediate | 2 | 2.0 | |
Multidrug resistance
95% of the isolates were found to be multidrug resistant. Six E. coli isolates were resistant to 7 antibiotics.
ESBL production
Out of 100 isolates of E. coli, 36 were detected as ESBL producers and 64 were non-ESBL producers.
Table 2 indicates the resistance pattern of clinical isolates against the antimicrobial agents in relation to ESBL production.
Table 2 Resistance pattern of clinical isolates from UTI (n=100) against antibioticsin relation to ESBL production.
| Clinical isolates from urinary | ||||||
|---|---|---|---|---|---|---|
| tract infection with E.coli | ||||||
| Antibiotic | (n=100) | |||||
| resistance | ESBL | Non ESBL | ||||
| pattern | producer | producer | ||||
| (n=36) | (n=64) | |||||
| n | % | n | % | |||
| ▪AMP 10 ug | ||||||
| Resistant | 36 | 100 | 59 | 92.2 | ||
| Sensitive | 0 | 0 | 3 | 4.7 | ||
| Intermediate | 0 | 0 | 2 | 3.1 | ||
| ▪SXT 25 ug | ||||||
| Resistant | 29 | 80.6 | 40 | 62.5 | ||
| Sensitive | 6 | 16.7 | 22 | 34.4 | ||
| Intermediate | 1 | 2.8 | 2 | 3.1 | ||
| ▪NA 30 ug | ||||||
| Resistant | 30 | 83.3 | 40 | 62.5 | ||
| Sensitive | 5 | 13.9 | 17 | 26.6 | ||
| Intermediate | 1 | 2.8 | 7 | 10.9 | ||
| ▪NOR 10 ug | ||||||
| Resistant | 26 | 72.2 | 33 | 51.6 | ||
| Sensitive | 9 | 25.0 | 28 | 43.8 | ||
| Intermediate | 1 | 2.8 | 3 | 4.7 | ||
| ▪CN 10 ug: | ||||||
| Resistant | 10 | 27.8 | 21 | 32.8 | ||
| Sensitive | 23 | 63.9 | 36 | 56.3 | ||
| Intermediate | 3 | 8.3 | 7 | 10.9 | ||
| ▪F 300 ug | ||||||
| Resistant | 6 | 16.7 | 10 | 15.6 | ||
| Sensitive | 23 | 63.9 | 42 | 65.6 | ||
| Intermediate | 7 | 19.4 | 12 | 18.8 | ||
| ▪KF 30 ug | ||||||
| Resistant | 36 | 100 | 57 | 89.1 | ||
| Sensitive | 0 | 0 | 4 | 6.3 | ||
| Intermediate | 0 | 0 | 3 | 4.7 | ||
| ▪IPM 10 ug | ||||||
| Resistant | 0 | 0 | 2 | 3.1 | ||
| Sensitive | 36 | 100 | 60 | 93.8 | ||
| Intermediate | 0 | 0 | 2 | 3.1 | ||
It is clear that resistance of isolates to each antibiotic is higher in case of ESBL producers than in case of non-ESBL producers except for Gentamicin and Imipenem. Resistances against Ampicillin were 100% and 92% for ESBL producers and non-ESBL producer’s respectively. Resistances to Imipenem were 0% and 3.1% for ESBL producers and non-ESBL producers respectively.
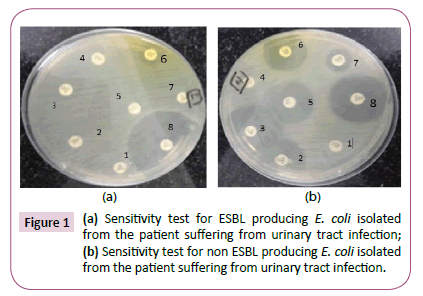
Figure 1 shows the differences in the resistance pattern of clinical isolates in relation to ESBL production.
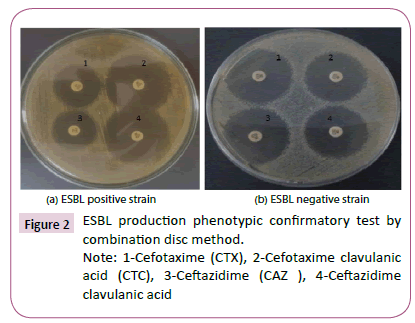
Figure 1a shows sensitivity test for ESBL producing E. coli isolate which show resistance to most antibiotics. In contrast, Figure 1b shows sensitivity test for non-ESBL producing E. coli isolate which show sensitivity to most antibiotics. Figure 2 shows ESBL production phenotypic confirmatory test by combination disc method.
Figure 1a shows ESBL positive strain. In contrast, Figure 1b shows ESBL negative strain.
Plasmid profiling
Plasmid profiling showed that out of 100 clinical isolates 68 were found to have one or more plasmids. Table 3 indicates the number of plasmids included in clinical isolates.
Table 3 Number and percentage of plasmids present in clinical isolates.
| No. of plasmids present | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| No. of including samples (total n=68) | 25 | 7 | 18 | 8 | 10 |
| % | 36.7 | 10.3 | 26.5 | 11.8 | 14.7 |
Number of plasmids was from 1 to 5. Out of 68 isolates, 25 (36.7%) contain 1 plasmid and 10 (14.7%) contain 5 plasmids.
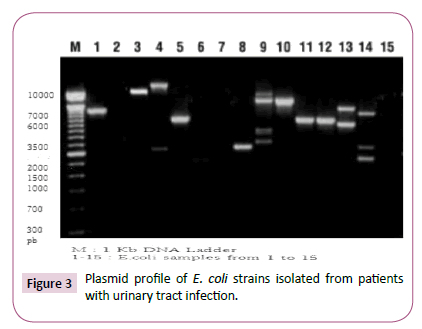
Figure 3 shows Plasmid profile of E. coli strains isolated from patients with urinary tract infection.
Plasmid profile analysis of E. coli strains revealed the presence of plasmids with various number and molecular weights in some samples and absence of plasmids in others.
Discussion
Urinary tract infection is one of the commonest bacterial infections [6]. It represents one of the most important causes of morbidity and is the second cause of hospital visits [14]. Urinary tract infection (UTI) forms the largest single group of hospitalacquired infection and accounts for about 40–50% of the total nosocomial infections. Enterobacteriaceae are the most frequent pathogens detected, causing 84.3% of UTI [15,16]. E. coli plays an important role in UTI [17].
The present study was done on 100 E. coli isolates recovered from 262 UTI urine specimens (38.2%). In a study [17], E. coli was the predominant uropathogen (54.6%) causing UTI. Similar results (E. coli 67%) were obtained by [18] as well as results obtained by; who stated that E. coli is still the most common cause of UTI [19] also reported 53% E. coli. Our results correspond with those of [20-23].
Antibiotic resistance is a major clinical problem in treating infections. The resistance of bacteria causing UTI to commonly prescribed antibiotics is increasing both in developing as well as in developed countries. Resistance has emerged even to more potent antimicrobial agents [24].
Our results showed that E. coli isolated were highly resistant to the so commonly used antibiotics: Ampicillin, 95%, Cephalothin 93%, Nalidixic acid 70%, Norfloxacin 59%, Trimethoprim Sulfamethoxazile 69% and Nitrofurantoin 16%. These results agreed with results of [25] who reported resistance rates similar to ours: Ampicillin 81.7%, cephalexin 92.7%, Nalidixic acid 78.9%, ciprofloxacin 49.5%, Norfloxacin 78.9%, co trimoxazole 54.1% and Nitrofurantoin 5.5%.
In the present study, 95% of the isolates were resistant to at least two antibiotics. Ninety five per cent of E. coli isolates were resistant to Ampicillin, a result that is in accordance with [6] (94%), [17] (88.7%) and [25] (81.7%). On the other hand, other studies showed lower results (60%) [26].
Extended-spectrum ß-lactamases are enzymes produced by a variety of organisms like enterobacteriaceae [27]. ESBL producing Enterobacteriaceae is an emerging public-health concern. Emergence of ESBLs in Gram negative bacteria reduces therapeutic options [19], because the ESBL-encoding isolates often co-express resistance to fluroquinolones, TMP-SMX, tetracycline and aminoglycoside, thus are often classified as multidrug-resistant pathogens [28]. Failure to detect these enzymes has contributed to their uncontrolled spread and therapeutic failure as well [29].
In the present study, ESBL isolated E. coli were all (100%) resistant to Ampicillin while the non-ESBL were 92.2% resistant to Ampicillin [18]. Revealed similar resistance results to the penicillin Amoxicillin (100% and 80.8%).
Seventy percent of our isolates were resistant to Nalidixic acid. Other studies revealed higher resistance rates as [30] and [17] who reported resistance rates of 89.2% and 87.2% respectively. [31] Recorded high resistance to Naidixix acid (61.2%). [25] recorded 57.7% though other studies showed lower results like the study done by [24]. It showed only 13% resistance to Naidixix acid.
In our ESBL group, resistance to Nalidixic acid was found to be 83.3% and 62.5% in non-ESBL E. coli studied which is in accordance with [18] who reported 81.8% and 59.6% in ESBL and non-ESBL repectively. Our results agreed also with those of [19] who reported 93% resistance in the ESBL group and 69% in the non-ESBL group.
We also reported 59% resistance to Norfloxacin which was high compared to results obtained [32] (11%) and [1] (11.2%) respectively. Our ESBL group showed resistance of 72.2% and the non- ESBL showed 51.6%. On the other hand, [18] reported 36.4% and 38.3% resistance to ciprofloxacin in ESBL and non- ESBL groups respectively. Meanwhile [19] showed 93% and 69% resistance in both ESBL and non-ESBL E. coli.
The resistance reported to quinolones and fluroquinolones could be attributed to the overuse of quinolones for treatment of UTI [32,33]. Another factor could be the generalized use of fluoroquinolones in animals feed (especially in poultry) and the subsequent transmission of resistance to strains from animals to humans [34].
Our study revealed 80.6% resistance to Trimethoprim/ Sulphamethoxazole in the ESBL, and 62.5% in the non-ESBL E. coli. Those results agreed with those of [18] who recorded 86.4% and 70.2% resistance to Cotrimoxazole in ESBL and non-ESBL studied E. coli. On the other hand, [19] revealed higher rate of resistance to Co-trimoxazole in ESBL (95%) but lower in non-ESBL (53%). On the contrary, results of [32] were lower 29% and 49% respectively.
Our research revealed that all ESBL E. coli (100%) were resistant to Cephalothin while the non ESBL ones showed 93% resistance.
The present study showed lower resistance to Gentamicin (31%), which is far from the result obtained by who reported 81%. Meanwhile Nitrofurantoin showed lower rates of resistance (16%).
Being a highly beta-lactamase stable carbapenem, with an unusual property of causing a post antibiotic effect on Gramnegative bacteria resistance to Imipenem was found to be very low (2%). This result agreed with that of [6] in Egypt and in India who detected showed no resistance (0%). On the other hand, higher resistances were shown by [35] and [1] were 43.3% and 32.5%.
Antimicrobial resistance (AMR) differs from a place to another depending on many factors. The increasing resistance to antibiotics is an inevitable result of the haphazard use of antibiotics by both physicians and lay people who have free access to over shelf antibiotics and their availability without even a prescription or a culture sensitivity testing. The improper antibiotic, dose and duration of administration are crucial factors for the emergence of AMR strains in community.
Besides using antibiotics in fish farms as well as the wide use in animal sector raised the resistance rates in animals and poultry, which in their turn transmit those resistant strains to human beings. The improper sanitation and hygiene is another cause of increased antimicrobial resistance. For antimicrobial resistance transmitted in hospitalized patients, the improper infection and prevention control measures is an important factor.
Much of the problem of antimicrobial resistance has been shown to be due to the presence of transferable plasmids encoding multidrug resistance and their dissemination among different enterobacterial species [7].
The results of the plasmid DNA analysis displayed that out of 100 strains; 68 contained plasmids responsible for resistance from 1 to 5 per isolate, and the other 32 strains did not include any plasmids. Molecular weights of the plasmids were between 1-33 kb. In a research conducted by Malkawi, the plasmids number was from 1 to 6 per isolate with sizes were from 1.5– 54 kb. The plasmid-free 32 isolates were found resistant to several antibiotics. The distribution of plasmids in all strains appeared that there was no specific plasmid profiling among examined isolates. This result confirmed that there was no linear correlation between the plasmids content and the antibiogram studies of the isolates. In this way, our findings were coherent with as mentioned previous studies [36-38].
Conclusion
The present research was conducted to study the resistance profile of clinical isolates from our local area against commonly prescribed antibiotics. Our results show very high degree of resistance to almost all antibiotics as compared to previously reported studies. The prevalence of ESBL producing E. coli is 36%. Multidrug resistance was found to be significantly higher in ESBL producer isolates as compared to non-ESBL producer isolates. In plasmid profiling, out of 100 strains; 68 contained plasmids and 32 were plasmid free. The plasmid-free 32 isolates were also found resistant to several antibiotics as well as those containing plasmids. The distribution of plasmids in all strains appeared that there was no specific plasmid profiling among examined isolates. The study showed the importance of continuous monitoring programming of multidrug resistance in our hospitals. It also showed the need for developing attempts to decrease the prevalence of ESBL producing organisms and the modification of guidelines for UTIs. ESBLs are clinically significant and patients infected with ESBL-producing Enterobacteriaceae experience a greater likelihood of poor outcome if they are treated with inappropriate antibiotics. The exception is uncomplicated urinary tract infections where a very high urinary concentration of β-lactam antibiotics can be achieved.
Recommendations
From our study findings it is highly recommended to have urgent surveillance of not only ESBL producers but also other antibiotic resistances in large number of cohorts in order to obtain prevalence of AMR producers. Surveillance should cover all sectors: human, animal and environmental health. Infection prevention and control measures should be applied vigorously and properly in all healthcare facilities. Application of Antimicrobial Stewardship program is mandatory in all healthcare facilities to increase the level of knowledge of physicians by education as regards antibiotics and their wise use and prescription. Over the shelf antibiotics prevention should be mandated by strict applied rules. The presence of at least a clinical pharmacist beside an infectious disease specialist and an efficient microbiologist are mandatory in each hospital for an effective Antimicrobial Stewardship Program.
Controlling the emergence and spread of ESBL organisms involves a combination of controlling antibiotic use and strict adherence to hospital infection control measures. Restriction of one class of antibiotics can lead to increased use of another class with an accompanying increase in resistance rates. If an isolate is confirmed as an ESBL-producer by the CLSI-recommended phenotypic confirmatory test procedure:
- All penicillins, cephalosporins, and aztreonam should be reported as resistant.
-This list does not include the cephamycins (cefotetan and cefoxitin), which should be reported according to their routine test results.
For treatment of ESBL producing bacteria:
-The cephamycins (e.g. cefoxitin, cefotetan) are structurally more stable than other cephalosporins to ESBL.
-Cefepime, a fourth-generation cephalosporin, is active against most ESBL-producing organisms. However, it should be given at high dose (≥ 2 g every 12 hours) usually in combination with other active agents (amino glycosides, fluoroquinolones).
- Currently, carbapenems are generally regarded as the preferred agent for treatment of infections due to ESBL-producing organisms.
-ESBLs are usually inhibited by β-lactamase inhibitors, such as clavulanic acid, sulbactam or tazobactam.
-Infection Control Issues: Several infection control factors should be emphasized when faced with combating the spread of ESBLproducing organisms. These issues include such things as isolation precautions, environmental decontamination and antibacterial usage patterns.
References
- Sabir S, Anjum AA, Ijaz T (2014) Isolation and antibiotic susceptibility of E. coli from urinary tract infections in a tertiary care hospital.
- WHO(2011) International Conference on Prevention & Infection Control (ICPIC).WHO, Geneva, Switzerland.
- Abhilash (2010) Epidemiology and outcome of bacteremia caused by extended spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiellaspp in a tertiary care teaching hospital in south India.J Assoc Physicians India 58: 13: 7.
- Momtaz H (2013) Serogroups virulence genes and antibiotic resistance in Shiga toxin-producing Escherichia coli isolated from diarrheic and non-diarrheic pediatric patients in Iran.Gut Pathog 5: 39.
- Taneja N, Rao P, Arora J, Ashok DA (2008) Occurrence of ESBL and Amp-C β-lactamases & susceptibility to newer antimicrobial agents in complicated UTI.Ind J Med Res 127: 85-88.
- Hassan S, Jamal SA, Kamal M (2011) Occurrence of multidrug resistant and esbl producing E.coli causing urinary tract infections.Australian Journal of Basic and Applied Sciences 7: 39-43.
- McPherson P, Gealt M (1986) Isolation of indigenous wastewater bacterial strains capable of mobilizing plasmid pBR325 Appl Environ Microbiol 51: 904-909.
- Kalantar E, Soheili F, Salimi H, SoltanDallal MM (2011) Frequency, antimicrobial susceptibility and plasmid profiles of Escherichia coli pathotypes obtained from children with acute diarrhea. Jundishapur. J Microbiol 4: 23-28.
- Cheesbrough M (1989) Medical laboratory manual for tropical countries. Microbiology Cambridge, Great Britain. Pp: 248-263.
- Holt JG, Krieg NR (1994) Bergey'smanual of determinative bacteriology(9th edn.), The Williams & Wilkins Co, Baltimore.
- Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J ClinPathol. 45: 493-496.
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Robins-Browne RM, Bordun AM, Tauschek M (2004) Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg Infect Dis 10: 1797-805.
- Kolawale AS, Kolawole OM, Kandaki-Olukemi YT, Babatunde SK, Durowade KA, et al. (2009) Prevalence of urinary tract infections (UTI) among patients attending DalhatuAraf Specialist Hospital LafiaNasarawa State Nigeria Int J Med MedSci 1: 163-167.
- Wada T, Iwamoto T (2009) Allelic diversity of variable number of tandem repeats provides phylogenetic clues regarding the Mycobacterium tuberculosis Beijing family. Infect Genet Evol 9: 921-926.
- Gales (1998) Activity and spectrum of 22 antimicrobial agents tested against urinary tract infection pathogens in hospitalized patients in Latin America: Report from the second year of the SENTRY Antimicrobial Surveillance Program.J AntimicrobChemother 45: 295-303.
- Manikandan C, Amsath A (2014) Antibiotic susceptibility pattern of Escherichia coli isolated from urine samples in PattukkottaiTamilnadu.International Journal of Current Microbiology and Applied Sciences.
- Islam MS, Yusuf MA (2014) Extended spectrum beta lactamase producing uropathogenicE.coli infection in Dhaka Bangladesh.Journal 0f Bacterioology Research 7: 1-7.
- Farzana R, Shamsuzzaman SM, Mamun KZ, Shears P (2013) Antimicrobial susceptibility pattern of extended spectrum b-lactamase producing gram-negative bacteria isolated from wound and urine in a tertiary care hospital Dhaka city, Bangladesh.Southeast Asian J Trop Med Public Health 44: 96-103.
- Jakobsen L, Garneau P, Bruant G, Harel J, Olsen SS, et al. (2012) Is Escherichia coli urinary tract infection a zoonosis? Proof of direct link with production animals and meat. Eur J ClinMicrobiol Infect Dis 31: 1121-1129.
- Agarwal J, Srivastava S, Singh M (2012) Pathogenomics of uropathogenic Escherichia coli. Indian J Med Microbiol 30: 141-149.
- Gautam (2013) Biotechnology International. Antibiotic resistance and its detection: role of specific proteins of multidrug resistance (mdr) strains.
- Ghazi K, Sajjad A, Sadia A (2013) Frequency of uropathogens in different gender and age groups. GJMS 11: 20-23.
- Thiraviam M, Yadesa D, Adugna T (2014) Antibiotic resistant pattern of urinary tract infection causing Escherichia coli isolated from diabetic mellitus and non-diabetic mellitus patients with special reference to Rifampicin resistance.Int J CurrMicrobiol App Sci 3: 668-674.
- Sharma AR, Bhatta DR, Shrestha J, Banjara MR (2013) Antimicrobial susceptibility pattern of escherichia coli isolated from urinary tract infected patients attending bir hospital.Nepal Journal of Science and Technology 14: 177-184.
- Arsalan H, Azap OK, Ergonul O, Timurkaynak F (2005) Risk factors for ciprofloxacin resistance among Escherichia coli strains isolated from community acquired urinary tract infection in Turkey. J AntimicrobChemother 56: 914-918.
- Bradford (2001) Extended spectrum beta-lactamases in the 21st century characterization, epidemiology and detection of this important resistance threat.ClinMicrobiol Rev 14: 933-951.
- Paterson DL, Bonomo RA (2006) Extended-spectrum beta-lactamases: A clinical update.ClinMicrobiol Rev 18: 657-686.
- Thomson KS, Moland ES (2001) Cefepime, piperacillin-tazobactam, and the inoculum effect in tests with extended-spectrum β-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother 45: 3548-3554.
- Kukanur (2015) Co-Relation between virulence factors and antibiotic resistance of E. coli with special reference to uropathogenic E. coli.IOSR Journal of Dental and Medical Sciences 14: 15-21.
- Nimri LF, Azaizeh BA (2012) First report of multidrug-resistant ESBLproducing urinaryEscherichia coli in Jordan.British Microbiology Research Journal 2: 71-81.
- Shakya (2013) Comparative metagenomic and rRNA microbial diversity characterization using archaeal and bacterial synthetic communities. Environ Microbiol 15: 1882-1899.
- Saleh AA, Ahmed SS, Ahmed M, Sattar ANI, Miah RA, et al. (2009) Changing trends in uropathogens and their antimicrobial sensitivity pattern. Bangladesh J Med Microbiol 3: 9-12.
- Miller LG, Tang AW (2004) Treatment of uncomplicated urinary tract infections in an era of increasing antimicrobial resistance.Mayo ClinProc 79: 1048-1053.
- Jafri SA, Qasim M, MasoudMS (2014) Antibiotic resistance of E. coli isolates from urine samples of urinary tract infection (UTI) patients in Pakistan.Bioinformation 10: 419-422.
- Walia S, Madhavan T, Williamson T, Kaiser A, Tewari R, et al. (1988) Protein patterns, serotyping and plasmid DNA profiles in the epidemiologic fingerprinting ofPseudomonas aeruginosa. Eur J ClinMicrobiol Infect Dis 7: 248.
- Pardesi KR, Yavankar SP, Chopade BA (2007) Plasmid distribution and antimicrobial susceptibility patterns of Acinetobactergenospecies from healthy skin of a tribal population in Western India. Indian J Med Res 125: 79-88.
- Farshad S, Ranjbar R, Japoni A, Hosseini M, Anvarinejad M, et al. (2012) Microbial susceptibility, virulence factors and plasmid profiles of uropathogenic Escherichia coli strains isolated from children in Jahrom Iran Arch Iran Med 15: 312-316.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences