ISSN : 2348-1927
Annals of Biological Sciences
Symptomatology and Molecular Variations of Graminicolous Downy Mildew Pathogens in Hybrids and Inbreds of Maize
Radhajeyalakshmi R1*, Harish S2 and Karthikeyan G2
1Department of Plant Research, Maize Research Station Tamil Nadu Agricultural University, Vagarai 624613, India
2Department of Plant Pathology, Tamil Nadu Agricultural University, Coimbatore 641003, India
- *Corresponding Author:
- Radhajeyalakshmi R
Department of Plant Research,
Maize Research Station Tamil Nadu Agricultural University,
Vagarai 624613,
India,
Tel: 7373249511;
E-mail: radhajeyalakshmi@hotmail.com
Received date: May 15, 2023, Manuscript No. ABS-23-16632; Editor assigned date: May 18, 2022, PreQC No. ABS-23-16632 (PQ); Reviewed date: June 02, 2023, QC No. ABS-23-16632; Revised date: July 25, 2023, Manuscript No. ABS-23-16632 (R); Published date: August 01, 2023, DOI: 10.36648/2348-1927.11.5.102
Citation: Radhajeyalakshmi R, Harish S, Karthikeyan G (2023) Symptomatology and Molecular Variations of Graminicolous Downy Mildew Pathogens in Hybrids and Inbreds of Maize. Ann Bio Sci Vol:11 No:5
Abstract
This study was performed to identify Peronosclerospora species found in Tamil Nadu based on sequence analysis of the COX-2 gene. In addition, sequence data in total, 25 in breeds infected with isolates of Peronosclerospora were investigated in this study. They were obtained from gene bank accessions of maize research station, Vagarai, Tamil Nadu and raised in Kharif and Rabi seasons. Both infected seeds and leaves were tested for the DNA analysis. Sequence analysis of COX-2 and phylogenetic inference were performed on 5 isolates. A set of primers developed in this study, PCOX2F and PCOX2R, was used for PCR amplification. Phylogenetic analyses showed that 5 isolates were divided into three groups. Maize crop is infected with 3 species of Pernosclerospora in two different growing seasons.
Keywords
Peronosclerospora; Inbreds; Downy mildew; PCR amplification
Introduction
Graminicolous Downy Mildews (GDMs) are diseases caused by members of the Peronosporaceae (Oomycota, Peronosporales) infecting cereal crops. GDM pathogens are obligate, biotrophic parasites, which are found to infect cultivated and wild cereals and other grasses in the Poaceae family [1]. The most destructive GDM pathogens reside in the regions of world found to cause significant crop losses of 60%–100% of staple food and forage crops viz., maize (Zea mays), pearl millet (Pennisetum glaucum), sorghum (Sorghum spp.) and sugarcane (Saccharum spp.) [2].
Downy mildew caused by the obligate oomycete Peronosclerospora genus, is one of the most devastating disease affecting maize (Zea mays L.) and can occur at any stage of maize development from seedling to harvest. The disease primarily infects its host from seedling emergence, until one month after planting. The Peronosclerospora species are known to attack members of the Poaceae in the tribes Andropogoneae, Maydeae and Paniceae [3]. The percentage of maize production areas with reported economic losses worldwide to downy mildew is 30%, both in tropical lowland maize and in subtropical, mid altitude, transition zone and highland maize [4]. In several maize growing countries including India, yield losses can reach 50%-100% for susceptible cultivars. Maize has been reported as host of eight Peronosclerospora species: P. heteropogoni, P. maydis, P. miscanthi, P. philippinensis, P. sacchari, P. sorghi, P. spontanea and P. eriochloae [5]. Identification of Peronosclerospora species has traditionally been based on host genus and morphological characteristics with limited discrimination ability. Consequently, identification of species is often unreliable [6]. Sporangial dimensions are a main characteristic element for species delimitation in Peronosclerospora, especially for those species without a known sexual stage and may be influenced by climatic condition, host species, and variety [7].
Among the detection methods of downy mildew species, microsatellites or SSRs have been recognized as one of the most informative type of markers for molecular ecology. SSRs are short oligonucleotides tandemly repeated motifs of one to six bases which occur frequently and randomly in all eukaryotic genomes, although their frequency varies significantly among different organisms [8]. The reliability of ARDRA in resolving species differences were well documented by Lukeman, et al. The tool ARDRA can be utilized to know the variation of noncoding regions, like the Internal Transcribed Spacer (ITS) and coding rDNA regions like the D1 and D2 of the 28S rDNA gene [9]. We will be employed these molecular techniques to survey the genetic variation among Tamil Nadu maize downy mildew isolates using COX2 primers obtained from known sequences of Pernospora sps. The objective of this study was to elucidate, whether the differences in disease response are more likely due to environmental factors, or due to genetic variation of downy mildew isolates among locations. The knowledge obtained from this research can be utilized to design effective strategies for maize downy mildew control.
Materials and Methods
In vivo screening of Peronosclerospora infected inbreds in Kharif and Rabi seasons
The present investigation was carried out during 2021–2022 at maize research station, Vagarai, Tamil Nadu, agricultural university, which is located at 10.58⁰ N latitude, 77.57⁰ E longitude with an altitude of 254 MSL in the Dindigul region of Tamil Nadu state, India. The test entries were raised in 2/3 plot size with two replications maintaining spacing of 60 cm between rows and 20 cm between plants. Susceptible check was sown on the borders of each plot. Observations on disease severity were recorded from 45 days after sowing [10]. Disease scoring was done until cob development stage. Rating scale observations from seedling to flowering are recorded on percentage basis, i.e. diseased plants as percent of total plant stand. Symptoms were categorized and grouped.
Molecular identification
DNA extraction from the type specimens of P. sorghi was done as described in Telle, et al., and also PCR was done according to that publication. In this study, a set of primer was developed from available oomycete sequences of cytochrome C oxidase subunit II (COX2) that were used for PCR amplification, namely PCOX2F (TCCAGCAA CTCCAGTTATGG) and PCOX2R (ACCTGGACAAGCAT CTAATT). This primer pair produces an amplicon of 529 bp. Sequences were compared with submitted sequences of NCBI.
Results and Discussion
The Graminicolous Downy Mildews (GDM) pathogens reside, are destructible to cause significant crop losses (60%–100%) of staple food and forage crops such as maize (Zea mays), pearl millet (Pennisetum glaucum), sorghum (Sorghum spp.), and sugarcane (Saccharum spp.) [11].
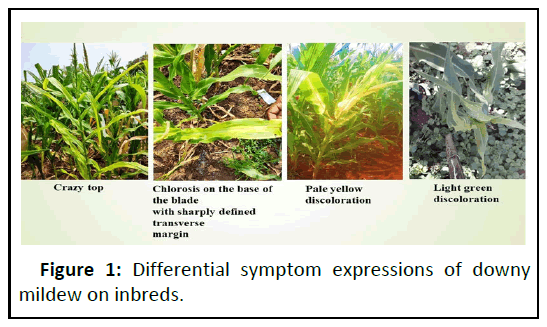
Java downy mildew caused by Peronosclerospora maydis is one of the three most devastating downy mildew pathogens of maize [12]. Crop damages of 40%–100% have been recorded in Indonesia. Disease symptoms varies from severe chlorosis in the upper leaves, stunting, deformation and lodging; infections by Peronosclerospora maydis may lead to death in susceptible maize varieties [13]. The pathogen is widely distributed in the tropics of Australia, China, India, Indonesia, Jamaica, Taiwan, Thailand, and Venezuela (Figure 1).
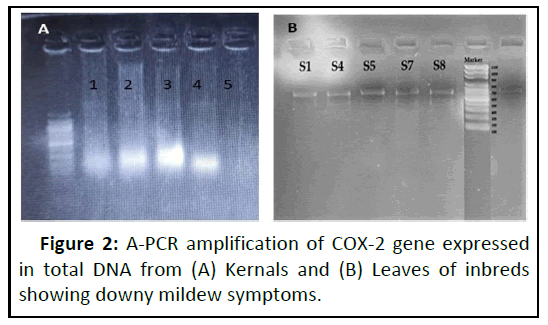
Based on the symptoms and partial COX2 sequencing, the most predominant species infecting maize is found to be Peronosclerospora , Indian isolates of 700 bp size and they have grouped distinctly compared with known sequences of P. sorghi and P. mayis (Figures 2 and 3). The sequences were identical with P. sorghi. These isolates can able to cause light green and yellow discolorations on the leaves. Severe chlorosis represents Pernosclerospora maydis, Scleropthora macrospora can be able to cause crazy top on the tassel (Figure 1). Dwarfing and cholosis represents P. sorghi and chlorosis streaks thorough out the leaf blade represents P. philippinensis [14-16]. The disease can be able to infect the crop during Kharif and Rabi seasons prevailing in these tracts. The COX-2 expressions in kernals reveals the seed borne nature of these pathogens.
Morphology based identification of downy mildews has many constraints, molecular identification is crucial and vital. Cytochrome C Oxidase subunit II (COX-2) has been found to be well-suited tool to differentiate species of oomycetes including Peronosclerospora [17]. Telle, et al., revealed that specimens of Peronosclerospora from maize and sorghum formed four distinct phylogenetic groups different from the known Peronosclerospora species included in the phylogeny [18]. Apart from the airborne nature of maize downy mildew, sorghum and maize downy mildew (P. sorghi) and pearl millet downy mildew (Sclerospora graminicola) were also reported both as seed and soilborne pathogens [19]. Of Peronosclerospora species that attack maize, P. sorghi is the only well studied species. This study will help to identify the genetic diversity of downy mildews infecting maize. Hence, there may be chances of mixed infections in maize, when they grown in Kharif and Rabi seasons in this tract (Table 1) [20].
| Inbreds | Dwarf | Interveinal chlorosis | Downy mildew | Crazy top | Species |
|---|---|---|---|---|---|
| VIM 026 | + | + | - | - | P. sorghi |
| VIM 027 | + | - | - | - | P. maydis |
| VIM 029 | + | + | - | - | P. maydis |
| VIM 045B | + | + | - | - | P. sorghi |
| VIM 110 | + | + | - | - | P. sorghi |
| VIM 131 | - | + | - | - | P. philippinensis |
| VIM 239 | - | + | - | - | P. philippinensis |
| VIM 419 | - | + | - | - | P. philippinensis |
| CoHM 6 | - | + | - | + | P. philippinensis and S. macrospora |
Table 1: Inbreds showing symptoms of downy mildews.
Conclusion
Based on the symptoms and partial COX-2 sequencing, the most predominant species infecting maize is found to be Peronosclerospora, Indian isolates. Among the isolates, dwarfing and cholosis will be due to P. sorghi and found to be predominant in both the crop growing seasons and rarely maize is found to be infected with Scleropthora macrospora causing crazy top, chlorosis streaks thorough out the leaf blade will be due to P. philippinensis. The diseases can be able to infect the crop during Kharif and Rabi seasons prevailing in these tracts. The COX-2 expressions in kernals reveals the seed borne nature of P. sorghi.
Acknowledgement
The author is thankful to Dr. Lakshmi Narayanan and Dr. K.R.V Sathyasheela for providing maize inbreds for field testing and screening in two seasons at maize research station, Vagarai, Tamil Nadu, India.
References
- Kubicke QB, Kenneth RG (1984) Peronosclerospora globosa, a new downy mildew of Gramineae, attacking cup grass in Texas. Phytopathol. 74:792
- Telle S, Shivas RG, Ryley MJ, Thines M (2011) Molecular phylogenetic analysis of Peronosclerospora (Oomycetes) reveals cryptic species and genetically distinct species parasitic to maize. Eur J Plant Pathol 130:521-528
- Weston WH (1921) Another conidial Sclerospora of Philippine maize. J Agric Res 20:669–684
- Kimigafukuro T (1979) Effect of temperature on conidial size of Sclerospora maydis, S. philippinensis and S. sorghi. JARQ 13:76-77
- Bock CH, Jeger MJ, Mughogho LK, Cardwell KF, Mtisi E, et al. (2000) Variability of Peronosclerospora sorghi isolates from different geographic locations and hosts in Africa. Mycol Res 104:61-68
- Selkoe KA, Toonen RJ (2006) Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol Lett 9:615-629
[Crossref] [Google Scholar] [PubMed]
- Lukman R, Afifuddin A, Lubberstedt T (2013) Unraveling the genetic diversity of maize downy mildew in Indonesia. J Plant Pathol Microb 4:162
- Choi YJ, Beakes G, Glockling S, Kruse J, Nam B, et al. (2015) Towards a universal barcode of oomycetes–a comparison of the COX-1 and COX-2 loci. Mol Ecol Resour 15:1275-1288
- Lukman R, Afifuddin A, Lubberstedt T (2016) Tracing the signature of Peronosclerospora maydis in maize seeds. Australas Plant Pathol 45:73-82
- Crouch JA, Davis WJ, Shishkoff N, Castroagudin VL, Martin F, et al. (2022) Peronosporaceae species causing downy mildew diseases of Poaceae, including nomenclature revisions and diagnostic resources. Fungal Syst Evol 9:43-86
[Crossref] [Google Scholar] [PubMed]
- Jeffers D, Cordova H, Vasal S, Srinivasan G, Beck D, et al. (2000) Status in breeding for resistance to maize diseases at CIMMYT. InProceedings of 7th Asian Regional Maize Workshop, Los Banos, Philippines 257-266
- Kumar A, Manga VK, Gour HN, Purohit AK (2013) Pearl millet downy mildew: Challenges and prospects. Ann Rev Plant Pathol 5:139
- Leu L (1973) Effects of temperature on conidial size and sporulation of Sclerospora sacchari. Plant Prot Bull 15:106–115
- Li R, Han Y, Zhang Q, Chang G, Han Y, Li X, et al. (2020) Transcriptome profiling analysis reveals co-regulation of hormone pathways in foxtail millet during Sclerospora graminicola infection. Int J Mol Sci 21:1226
[Crossref] [Google Scholar] [PubMed]
- Perumal R, Nimmakayala P, Erattaimuthu SR, No EG, Reddy UK, et al. (2008) Simple sequence repeat markers useful for sorghum downy mildew (Peronosclerospora sorghi) and related species. BMC Genet 9:1-4
- Pudjiwati EH, Kuswanto K, Basuki N, Sugiharto AN (2013) Path analysis of some leaf characters related to downy mildew resistance in maize. J Agric Sci 35:167-173
- Putnam ML (2007) Brown stripe downy mildew (Sclerophthora rayssiae var. zeae) of maize. Plant Health Prog 8:34
- Rathore RS, Trivedi A, Mathur K (2002) Rajasthan downy mildew of maize: The problem and management perspectives. InProceedings of the 8th Asian regional maize workshop, Bangkok, Thailand 366-379
- Spencer MA, Dick MW (2002) Aspects of graminicolous downy mildew biology: Perspectives for tropical plant pathology and peronosporomycetes phylogeny. Trop Mycol Micromyce 2:63-81
- Thines M, Telle S, Choi YJ, Tan YP, Shivas RG (2015) Baobabopsis, a new genus of graminicolous downy mildews from tropical Australia, with an updated key to the genera of downy mildews. IMA fungus 6:483-491
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences