Strategies for the Production of Soluble Recombinant Proteins Using Escherichia Coli: A Review
1Center for Advanced Sensor Technology, University of Maryland Baltimore County, 1000 Hilltop Circle, Baltimore, Maryland 21250, USA
2Department of Chemical and Biological Engineering, Korea University, Anamdong, Seongbuk-gu, Seoul, Korea
3Center for Computational and Integrative Biology, Department of Biology, Rutgers, The State University of New Jersey, Camden, NJ, United Statesa
- *Corresponding Author:
- Chandrasekhar Gurramkonda
Center for Advanced Sensor Technology,
University of Maryland Baltimore County,
1000 Hilltop Circle, Baltimore, Maryland
21250, USA
E-mail: chskrg@gmail.com
Received date: August 29, 2018; Accepted date: October 27, 2018; Published date: November 05, 2018
Citation: Gurramkonda C, Gudi SK, Koritala BSC (2018) Strategies for the Production of Soluble Recombinant Proteins Using Escherichia Coli: A Review. J Mol Biol Biotech Vol.3 No.1:6
Abstract
Escherichia coli (E. coli) have been widely used genetic model for the production of recombinant proteins. Because of its simplicity in cloning and fast replication, scientists have chosen E. coli to generate higher yields of heterologous proteins. In recent years, the demand for recombinant proteins increased for biomedical applications from basic research to diagnostics. However, the abundance in the expression of soluble recombinant proteins in E. coli is challenging. More often, recombinant proteins misfold and form aggregates in the host. In this chapter, we propose the strategies for enhancing the soluble expression of disulfide bond proteins and non-disulfide bond proteins.
Keywords
E. coli; Soluble protein expression; Viral proteins; Antibodies; Fusion proteins and Chaperones
Introduction
From 1980s recombinant DNA technology has been widely used for the production of therapeutically valuable proteins using microbial host systems [1]. This technology has been rapidly emerging in the field of biotechnology, which prevents to sacrifice vast numbers of wild or domestic animals for producing therapeutic proteins. Federal regulatory agencies are widely accepted to use E. coli as a model to produce heterologous proteins [2,3]. The expression of foreign proteins in the microbial systems is challenging because these proteins generate metabolic burden to the host. Due to the foreignness, producing higher concentrations of soluble heterologous protein is a significant problem. Because these foreign proteins tend to misfold and form insoluble inclusions in the cytoplasm of the host [4,5].
Several viral and anti-viral protein antibodies significantly explored for controlling viral infections [6]. Recombinant protein production categorically grouped into enzymes, anticoagulants, scaffold proteins, antibodies, growth factors, interferons, and hormones. Several commercial host expression systems like prokaryotes (bacteria) and Eukaryotes (yeast, fungi, insects, mammalian cells, plants, and animals) have been used to produce heterologous recombinant proteins (Table 1).
Among them, E. coli has been widely reported for producing recombinant proteins. As mentioned earlier, recombinant protein production has challenges to control the inclusion body formation. To overcome this problem selecting specific recombinant constructs, which are compatible with particular hosts are essential. Additionally, maintaining plasmid copy number, inducer concentrations, growth medium, protein folding mechanisms, protein docking, purification strategies are helpful to enhance the final yield of the soluble recombinant protein [15-17].
Studies have shown that presence or absence of disulfide bonds strongly influences protein folding mechanisms to strengthen the expression of the soluble recombinant protein. Mispairing of disulfide bonds leads to protein denaturation, which forms inclusion bodies. In parallel, increase in the number of cysteine residues leads to misfolding of recombinant proteins. Such circumstances, foldases or chaperones assist the protein folding process and minimizes the misfolding of recombinant proteins [18,19]. Prevention of inclusion bodies is cost-effective and challenging for scientists across the globe. There are several standardized refolding techniques were established to recover the biologically active form of the recombinant proteins [20]. In the current review, we focus on how to enhance the solubility of recombinant protein expression in E. coli.
Fusion Partners and Chaperones
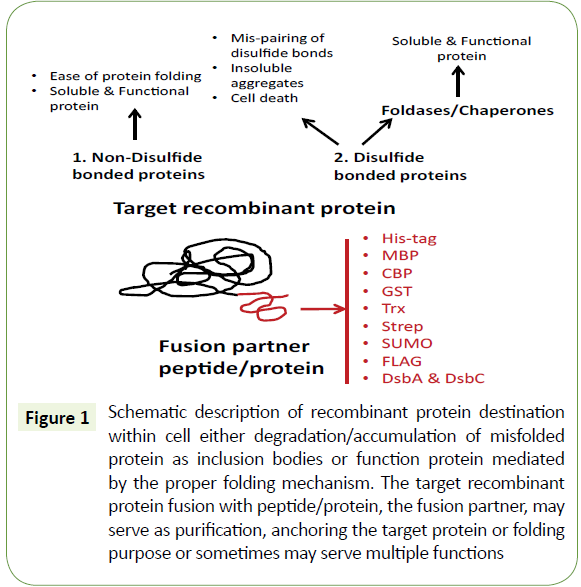
Biopharmaceutical industries are widely using gene fusion methods for enhancing the solubility of recombinant proteins. Biomedical research has shown that fusion partners are playing an essential role in increasing the stability of blood circulation, controlling cytotoxicity and methods for targeted drug delivery, etc. [21]. The role of fusion partners in the expression of recombinant proteins are categorized mainly based on its function. Affinity tags and solubility enhancing tags are mostly used for protein purification and protein folding respectively. Affinity tags are often a short stretch of amino acid sequence that shows a strong affinity towards the chromatography matrix immobilized ligand (Figure 1).
Figure 1: Schematic description of recombinant protein destination within cell either degradation/accumulation of misfolded protein as inclusion bodies or function protein mediated by the proper folding mechanism. The target recombinant protein fusion with peptide/protein, the fusion partner, may serve as purification, anchoring the target protein or folding purpose or sometimes may serve multiple functions.
The affinity tag fusion to the recombinant proteins work based on the single step purification without prior knowledge of any known target proteins. Recent advances in affinity tags enhance the purification rate. However, scientists are still looking for unique affinity tags to improve protein purification methods. Among them, different lengths of polyhistidine affinity tags were used extensively for recombinant protein production using plasmid expression system (pET vectors). The polyhistidine fusion forms readily co-ordination bond with polycationic metal ions immobilized matrix material, which offers single-step affinity purification. In addition to polyhistidine tags, most of the pET vectors were customized with tags of N-utilization substance A, thioredoxin, DsbA, DsbC and the peptidyl-prolyl cis-trans isomerase for enhancing and purifying the soluble recombinant proteins. Furthermore self-cleaving affinity tags like inteins and AKT protein kinase tags were used in purification of recombinant proteins. The other fusions tags, such as N-terminal glutathione S-transferase (GST) was constructed to pGEX plasmid expression system, which was also used for the purification using glutathione sepharose affinity column. However, fusion proteins crosslink to the resin often leads to inclusion bodies, dimerization of GST and it is a major drawback.
Additionally, tags of Maltose Binding Protein (MBP) often used for increasing the solubility of recombinant proteins. MBP tags can also serve as an affinity tag for protein purification. Recently, a study has shown that histidine maltose binding protein fusion tags enhances the soluble recombinant proteins. pMAL plasmid expression system is commercially available for cytoplasmic or periplasmic protein production. Amylose crosslinked resins were used for affinity purification of fusion proteins. The calmodulin binding peptide (CBP) fusion partners linked at C or N -terminal of targeted proteins. This purification system influenced by the presence of Ca2+ that affects the calmodulin conformational changes to release their fusion partners. Fusion proteins of the streptavidin-binding peptide are affinity purified by streptavidin immobilized resins, and fusion partners are further eluted with biotin solution.
The SUMO (small ubiquitin-like modifier) family proteins are fused to recombinant protein and form a covalent bond with lysine residues, and SUMO proteases later remove the affinity tags. The eight amino acid short peptide, FLAG, a hydrophilic short amino acid tag has been extensively used for immunoprecipitation and selectively captures fusion protein with the specific antibody. The affinity tags are not permanent domains to the target recombinant proteins, and their selective removal is often recommended. The list of fusion affinity tags used for recombinant protein purification listed in Table 2. Affinity purification tags are designed with target recombinant protein by having proteolytic cleavable recognition sequences between the fused partners.
Chaperone Based Solubility Enrichments
Chaperone is a significant component of proteostasis network that possesses the highly conserved function of preventing protein misfolding and aggregation. In E. coli, protein folding is controlled by four different chaperone systems. Among them, GroELS chaperonin system encapsulate client proteins and providing an isolated environment to achieve the state of native protein folding [26]. Another chaperone system in E. coli is more likely to Hsp 70 system of eukaryotes called as KJE system. This system has combination of three different chaperones DnaJ, DnaK and GrpE [27]. The major role of the KJE system in proteostasis is to unfold misfolded proteins, thereby giving another chance for unfolded proteins to refold. A combination of KJE with ClpB has been considered as disaggregation system (B+KJE) involved in protecting E. coli from heat shock [28]. Trigger factor has been excluded from other chaperone systems, which is engaged in preventing aggregation of translating polypeptides in E. coli [28].
Several studies have shown that co-expression of E. coli chaperones enhances the production of recombinant proteins. Co-expression of KJE chaperone components increases the solubility and production of recombinant proteins includes enzymes, antibodies, etc. [29,30]. In parallel, increasing solubility of heterologous proteins was reported with co-expression of other bacterial chaperones includes GroELS, ClpB and trigger factor. Even though there is a more significant advantage in coexpression of molecular chaperones with recombinant proteins, few studies have shown the negatively impact the co-expression of chaperones on quality and quantity of specific heterologous proteins [31].
The Past and Present Strategies of Soluble Expression of Recombinant Proteins
The foreignness and excessive production of recombinant protein in the host change the pH, osmolarity, redox potentials in the cytoplasm, which further causes self-aggregation of recombinant proteins in the host. The primary challenge for scientists is to develop and tune-up the recombinant expression system to minimize and prevent the recombinant protein aggregation and enhance the soluble protein production. There was no standard method for soluble expression of aggregation-prone recombinant proteins. Factors include a selection of appropriate host, codon optimization, designing vector constructs, expression conditions, and fusion or co-expression partners such as fold ases and molecular chaperones that interact with the nascent polypeptide to achieve the functional tertiary structure of recombinant proteins.
The trial and error of various plasmid designs constructs and microscale expression studies often provide valuable information for soluble expression. Recombinant protein aggregation as inclusion formation is influenced by improper protein folding and mispairing of disulfide bond formation. The recombinant target protein with increased cysteine residues often requires the oxidative cytoplasmic environment, co-expression of foldase/ molecular chaperones, cofactors, lower temperature for proper post-translational modification of recombinant protein to increase their solubility. The extensive track record of recombinant protein production in the literature can be divided into strategies before and later inclusion bodies formation (Table 3). The in-vitro solubilization and refolding is not practical industrial application due to its lesser recovery and cost. This will suggest the soluble protein expression is the most desirable process as a potential alternative to inclusion bodies production.
The other factors that determine the solubility of proteins include 1) amino acid sequence and buffer composition; 2) functional groups that determine the net charge; 3) Iso-electric point of the target protein; 4) turn forming residues; 5) low tRNA availability; 6) mRNA secondary structure, etc.
Multiple Disulfide Bonds and Strategies for Avoiding the Inclusion Bodies
The structural conformation of the recombinant protein determines the quantity of soluble functional protein. Formation of disulfide bonds between inters- and the intra-polypeptide chain is an oxidative process. For this process, the cytoplasmic environment of E. coli should maintain an oxidative state. Due to the production of heterologous proteins, cells may not preserve the oxidative rich environment, which impacts the formation of stable disulfide bonds. At this stage, the cell uses its molecular machinery, which includes catalyzing enzymes, chaperones, and foldases to obtain the structural conformation of recombinant proteins by avoiding protein misfolding and aggregation. The naturally occurring non-disulfide bond proteins include polymerases, alcohol/lactate dehydrogenases, protein G, lysozyme, Calmodulin, casein, etc.
In eukaryotes, cellular organelles were compartmentalized with oxidative rich microenvironment, which mediates the formation of disulfide bonds with the nascent polypeptides. In contrast, prokaryotes like E. coli do not possess compartmentalized cellular organelles to maintain oxidative rich microenvironments. However, periplasmic space substitutes to maintain the oxidative rich microenvironments for protein folding processes. In E. coli, cytoplasmic redox potentials are negatively maintained by thioredoxin (trxB) and glutathione-based system. Mutants of thioredoxin and glutathione strains have shown efficiency in synthesizing disulfide bonds with native polypeptides [46].
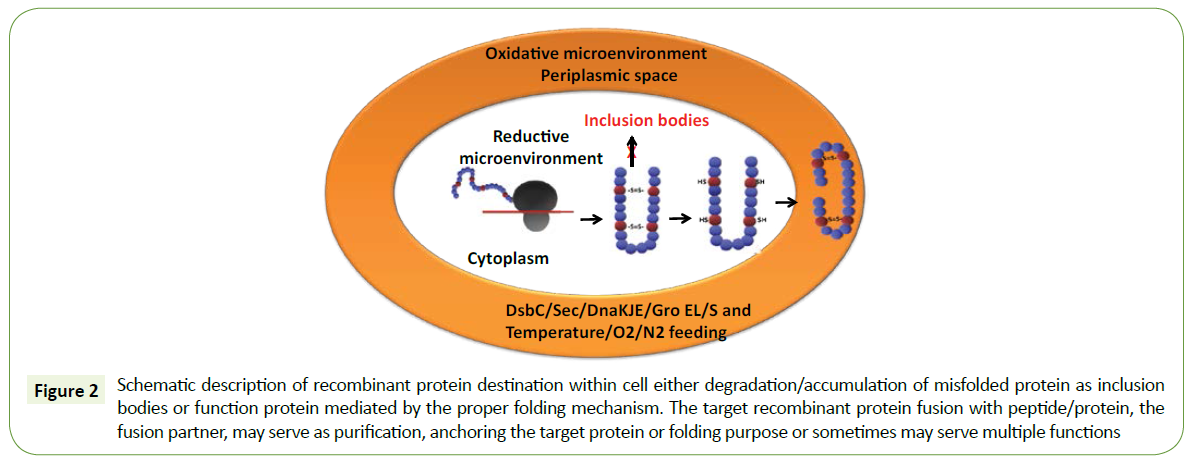
Protein folding of the nascent polypeptide chain in the cytoplasm is achieved by different environmental conditions of the host cell. The first mechanism is to direct periplasmic space by N-terminal signal peptide or Sec-dependent mechanism, and this oxidative microenvironment favors the correct disulfide bond formation and to achieve the native structural conformation. The second way to the nascent polypeptide chain in the cytoplasm is to create oxidative microenvironment through trxB and gor/gshA mutant E. coli strains, co-expression of protein disulfide isomerase DsbC and cytoplasmic chaperone (Figure 2).
Figure 2: Schematic description of recombinant protein destination within cell either degradation/accumulation of misfolded protein as inclusion bodies or function protein mediated by the proper folding mechanism. The target recombinant protein fusion with peptide/protein, the fusion partner, may serve as purification, anchoring the target protein or folding purpose or sometimes may serve multiple functions
The nascent polypeptide chain in the cytoplasm has several destinations to achieve the structural and functional protein. The first mechanism is to direct periplasmic space by N-terminal signal peptide or Sec-dependent mechanism, and this oxidative microenvironment favors the correct disulfide bond formation and to achieve the native structural conformation. The second way to the nascent polypeptide chain in the cytoplasm is to create oxidative microenvironment through trxB and gor/gshA mutant E. coli strains, co-expression of protein disulfide isomerase DsbC and cytoplasmic chaperone (Figure 2).
Production of Recombinant Viral Proteins, Antibodies, and Biopolymers Pha
Recombinant viral proteins
Viral proteins are expressed in various recombinant host strains using customized vectors to achieve stable expression system (Table 1). Elucidation for molecular interactions of individual viral proteins in replication of viruses is most important. The cultivation of microorganisms in large quantities is a highly complicated and an expensive process. An alternative approach is the production of individual functional viral proteins in recombinant hosts. The fully functional recombinant viral proteins should have the properties of the native protein, such as induction of protective response, antigenicity, immunogenicity, native viral protein structure, and self-assembly into multimeric form [9]. Classical examples, T7 RNA polymerase expression [47], recombinant adenovirus vectors by homologous recombination [48].
Recombinant antibodies
Production of antibodies by a traditional monoclonal antibody (mABs) technology is most useful for therapeutic and diagnostic applications, which is approved by FDA. The mABs production takes place in hamster ovary cells or mouse cell lines. The screening and selection of stable antibody-producing cell lines are the expensive, laborious and time-consuming process. The recombinant E. coli is a best and choice for antibody production. As discussed earlier, the major hurdle of antibody di-sulfide formation for improper protein folding could be minimized by trafficking the recombinant protein to an oxidative compartment of the periplasmic region or mutant strains of thioredoxin (trxB) and glutathione (gor or gshA). Immunoglobulin antibodies of fulllength or fragments such as the single-chain fragment of variable region Fv (scFv), Fc (fragment of crystallization) and Fab (short fragment of antibody binding) are classical examples [33,49,50].
Recombinant biopolymers PHA
The biopolymers, polyhydroxyalkanoates [51] are produced by most of the naturally occurring bacterial members as intracellular reserves of “c” source. The bacterial groups such as Ralstonia eutropha, Pseudomonas putida, Bacillus pumilus, Aeromonas ichthiosmia, etc., The PHA biopolymer has been used extensively in applications of bioplastics, drug delivery, and synthetic tissues. PHA synthases are metabolic enzymes play the viral catalytic role in the synthesis of PHA. PHA is an internal reserve, and several other regulatory genes can control the production of PHA. The recombinant technology is under evaluation and emerging field for the synthesis of biopolymers [52].
In conclusion, Successful recombinant protein production is indispensable for biomedical research. Although robust platforms are available, the limitations include the soluble protein production. In this review we presented the full scan of strategies for soluble recombinant protein production. We believe that this review will benefit the needs of the protein production community.
Funding
There was no funding for this review article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Johnson IS (1983) Human insulin from recombinant DNA technology. Science 219: 632-637.
- Olempska-Beer ZS, Merker RI, Ditto MD, DiNovi MJ (2006) Food-processing enzymes from recombinant microorganisms--a review. Regul Toxicol Pharmacol 45: 144-158.
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, et al. (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97: 5978-5983.
- de Marco A, Deuerling E, Mogk A, Tomoyasu T, Bukau B (2007) Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC biotechnol 7: 32.
- LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, et al. (1993) A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (N Y) 11: 187-193.
- Mazanec MB, Kaetzel CS, Lamm ME, Fletcher D, Nedrud JG (1992) Intracellular neutralization of virus by immunoglobulin A antibodies. Proc Natl Acad Sci USA 89: 6901-6905.
- Lebozec K, Jandrot-Perrus M, Avenard G, Favre-Bulle O, Billiald P (2018) Quality and cost assessment of a recombinant antibody fragment produced from mammalian, yeast and prokaryotic host cells: A case study prior to pharmaceutical development. N Biotechnol 44: 31-40.
- Chu C, Zhang W, Li J, Wan Y, Wang Z, et al. (2018) A single codon optimization enhances recombinant human tnf-alpha vaccine expression in Escherichia coli. Biomed Res Int 3025169.
- Gurramkonda C, Talha SM, Gudi SK, Gogineni VR, Surya Sambasiva Rao KR (2012) Fed-batch cultivation of Escherichia coli expressed designer hepatitis c virus diagnostic intermediate and its evaluation. Biotechnol Appl Biochem 59: 437-444.
- Moreno-Perez DA, Baquero LA, Bermudez M, Gomez-Munoz LA, Varela Y, et al. (2018) Easy and fast method for expression and native extraction of plasmodium vivax duffy binding protein fragments. Malar J 17: 76.
- Clare JJ, Rayment FB, Ballantine SP, Sreekrishna K, Romanos MA (1991) High-level expression of tetanus toxin fragment C in Pichia pastoris strains containing multiple tandem integrations of the gene. Biotechnology (N Y) 9: 455-460.
- Jiang X, Wang M, Graham DY, Estes MK (1992) Expression, self-assembly, and antigenicity of the norwalk virus capsid protein. J Virol 66: 6527-6532.
- Lai T, Yang Y, Ng SK (2013) Advances in mammalian cell line development technologies for recombinant protein production. Pharmaceuticals (Basel) 6: 579-603.
- Wigdorovitz A, Perez Filgueira DM, Robertson N, Carrillo C, Sadir AM, et al. (1999) Protection of mice against challenge with foot and mouth disease virus (FMDV) by immunization with foliar extracts from plants infected with recombinant tobacco mosaic virus expressing the FMDV structural protein VP1. Virology 264: 85-91.
- Hannig G, Makrides SC (1998) Strategies for optimizing heterologous protein expression in escherichia coli. Trends Biotechnol 16: 54-60.
- Hizi A, McGill C, Hughes SH (1988) Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci U S A 85: 1218-1222.
- Sorensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of escherichia coli. Microb Cell Fact 4: 1.
- Clark EDB (1998) Refolding of recombinant proteins. Curr Opin Biotechnol 9: 157-163.
- Sorensen HP, Mortensen KK (2005) Advanced genetic strategies for recombinant protein expression in escherichia coli. J Biotechnol 115: 113-128.
- Yamaguchi H, Miyazaki M (2014) Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies. Biomolecules 4: 235-251.
- Schmidt SR (2009) Fusion-proteins as biopharmaceuticals--applications and challenges. Curr Opin Drug Discov Devel 12: 284-295.
- Esposito D, Chatterjee DK (2006) Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol 17: 353-358.
- Guan KL, Dixon JE (1991) Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione s-transferase. Anal Biochem 192: 262-267.
- Kapust RB, Waugh DS (1999) Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 8: 1668-1674.
- Terpe K (2003) Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 60: 523-533.
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, et al. (2001) Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107: 223-233.
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, et al. (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683-689.
- Mogk A, Bukau B. (2004) Molecular chaperones: Structure of a protein disaggregase. Curr Biol 14: R78-80.
- Choi GH, Lee DH, Min WK, Cho YJ, Kweon DH, et al. (2004) Cloning, expression, and characterization of single-chain variable fragment antibody against mycotoxin deoxynivalenol in recombinant Escherichia coli. Protein Expr Purif 35: 84-92.
- Thomas JG, Baneyx F (1996) Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing heat-shock proteins. J Biol Chem 271: 11141-11147.
- Martinez-Alonso M, Garcia-Fruitos E, Ferrer-Miralles N, Rinas U, Villaverde A (2010) Side effects of chaperone gene co-expression in recombinant protein production. Microb Cell Fact 9: 64.
- Arechaga I, Miroux B, Karrasch S, Huijbregts R, de Kruijff B, et al. (2000) Characterisation of new intracellular membranes in escherichia coli accompanying large scale over-production of the b subunit of F(1)F(o) ATP synthase. FEBS lett 482: 215-219.
- Bessette PH, Aslund F, Beckwith J, Georgiou G (1999) Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci U S A 96: 13703-13708.
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852-1858.
- Lu X, He S, Zong H, Song J, Chen W, et al. (2016) Improved 1, 2, 4-butanetriol production from an engineered Escherichia coli by co-expression of different chaperone proteins. World J Microbiol Biotechnol 32: 149.
- Shuo-shuo C, Xue-zheng L, Ji-hong S (2011) Effects of co-expression of molecular chaperones on heterologous soluble expression of the cold-active lipase lip-948. Protein Expr Purif 77: 166-172.
- Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22: 346-353.
- Baneyx F, Mujacic M (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 22: 1399-1408.
- Cristobal S, de Gier JW, Nielsen H, von Heijne G (1999) Competition between sec- and tat-dependent protein translocation in Escherichia coli. EMBO J 18: 2982-2990.
- Baneyx F (1999) Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol 10: 411-421.
- Georgiou G, Valax P (1996) Expression of correctly folded proteins in Escherichia coli. Curr Opin Biotechnol 7: 190-197.
- Kopetzki E, Schumacher G, Buckel P (1989) Control of formation of active soluble or inactive insoluble baker's yeast alpha-glucosidase pi in Escherichia coli by induction and growth conditions. Mol Gen Genet 216: 149-155.
- Turner P, Holst O, Karlsson EN (2005) Optimized expression of soluble cyclomaltodextrinase of thermophilic origin in Escherichia coli by using a soluble fusion-tag and by tuning of inducer concentration. Protein Expr Purif 39: 54-60.
- Waugh DS (2005) Making the most of affinity tags. Trends Biotechnol 23: 316-320.
- Islam RS, Tisi D, Levy MS, Lye GJ (2007) Framework for the rapid optimization of soluble protein expression in Escherichia coli combining microscale experiments and statistical experimental design. Biotechnol Prog 23: 785-793.
- Prinz WA, Aslund F, Holmgren A, Beckwith J (1997) The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem 272: 15661-15667.
- Fuerst TR, Niles EG, Studier FW, Moss B (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A 83: 8122-8126.
- Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, et al. (1996) Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol 70: 4805-4810.
- Shibui T, Nagahari K (1992) Secretion of a functional fab fragment in Escherichia coli and the influence of culture conditions. Appl Microbiol Biotechnol 37: 352-357.
- Skerra A, Pfitzinger I, Pluckthun A (1991) The functional expression of antibody Fv fragments in Escherichia coli: Improved vectors and a generally applicable purification technique. Biotechnology (N Y) 9: 273-278.
- Davis JM, Arakawa T, Strickland TW, Yphantis DA (1987) Characterization of recombinant human erythropoietin produced in chinese hamster ovary cells. Biochemistry 26: 2633-2638.
- Taguchi S, Doi Y (2004) Evolution of polyhydroxyalkanoate (PHA) production system by "enzyme evolution": Successful case studies of directed evolution. Macromol Biosci 4: 146-156.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences