ISSN : 2573-4466
Insights in Enzyme Research
Purification and Characterization of Active Aggregates of an Organic Solvent Tolerant Lipase from Marinobacter sp. EMB5
R Hemamalini and Khare SK*
Enzyme and Microbial Biochemistry Lab, Department of Chemistry, Indian Institute of Technology, Hauz Khas, New Delhi, India
- *Corresponding Author:
- Khare SK
Enzyme and Microbial Biochemistry Lab, Department of Chemistry, Indian Institute of Technology, Hauz Khas, New Delhi, India.
Tel: +911126596533
Fax: +911126581102
E-mail: skhare@rocketmail.com;skkhare@chemistry.iitd.ac.in
Received Date: November 28, 2016; Accepted Date: December 09, 2016; Published Date: December 12, 2016
Citation: R Hemamalini, Khare SK. Purification and Characterization of Active Aggregates of an Organic Solvent Tolerant Lipase from Marinobacter sp. EMB5. Insights Enzyme Res. 2016, 1:1. doi: 10.21767/2573-4466.100003
Abstract
Background: Marinobacter is a halophilic bacterial genus and a bio-geochemically important group in sea water, is generally associated with the bioremediation of hydrocarbon-rich effluents released into the salt-rich environment by various anthropogenic activities. It is a well-known lipase producer-the NCBI Protein database shows nearly 520 Marinobacter lipases. In this context, its lipase and esterase are considered important. However, their enzymatic properties in purified form have been scantily studied. Since hydrocarbons are acted upon on solvent water interface, the stability and catalytic behavior of Marinobacter lipase in non-aqueous media needs to be more specifically investigated.
Aim: Marinobacter sp. EMB5 was isolated from Kozhikode, Kerala, India. It was found to be potent lipase producer. The aim of the present work was to obtain a purified preparation and undertake its detailed characterization with respect to above aspects.
Work: The crude Marinobacter sp. lipase was purified by Sephacryl S200 gel filtration chromatography. The lipase eluted as a single activity peak in void volume and had a very high molecular mass. Aggregation has not been reported in case of Marinobacter lipase, so far, although it is known in other lipases. The aggregated lipase was mixed with 2-propanol (70%, v/v) and disaggregated lipase was obtained as a single activity peak in Sephacryl S200 gel filtration chromatography corresponding to 82 kDa molecular mass. This monomeric species was less active than aggregated purified preparation. Hence, further characterization and studies were carried out on purified active aggregate lipase. It was stable in a wide range of organic solvents at high concentration up to 24 h. The organic solvent stability was further investigated by Far UV CD spectroscopy and fluorescence spectroscopy.
Conclusion: Due to the fact that Marinobacter sp. EMB5 lipase remained stable in organic solvents for prolonged incubation periods, it has potential to be used in industrial organic synthesis.
Keywords
Halophilic lipase; Solvent tolerant lipase; Marinobacter lipase; Active lipase aggregates; Haloalkaliphilic lipase
Introduction
Halophilic lipases are interesting for biotechnological applications as they retain their functionality even under high salt and lowwater conditions. Low-water conditions predominate within the halophilic cell, due to excessive accumulation of salts within the cytoplasm as an adaptation mechanism. In the industry, organic synthesis reactions carried out under solvent-rich conditions mimic such low-water conditions. Since the activity of halophilic lipases is not compromised even in presence of very high concentration of organic solvents, they are very promising candidates for carrying out lipase-catalyzed organic biotransformation [1-3].
Although a large number of halophilic lipases have been detected and listed on the NCBI’s protein database (https://www.ncbi.nlm. nih.gov/protein), very few have been purified and characterized in detail. These include Oceanobacillus rekensis PT-11 lipase [4], Haloarcula sp. G41 lipase [5], Idiomarina sp. W33 lipase [2] and LipBL from Marinobacter lipolyticus SM19 [6]. Moreover, not much is known about the correlation of their solvent stability with structural features. Structural studies further require purified form of lipase.
Purification of halophilic lipase, in general, poses problem for two reasons, (i) Requirement of incorporating the salt (necessary to maintain native structure of halophilic proteins) throughout the purification process. Otherwise, protein is denatured leading to loss of activity [7]. But the addition of salt disrupts the chromatographic procedure. (ii) Propensity of aggregation in lipase causes formation of aggregates during purification, making purification quite unpredictable.
Most halophilic lipases are also organic solvent tolerant owing to a similarity in the micro-environment created by the presence of organic solvents and high salt concentration. LipBL from Marinobacter lipolyticus displayed remarkable stability in presence of both hydrophobic as well as hydrophilic solvents, with maximum stability in presence of methanol [6]. The Idiomarina sp. W33 and Haloarcula sp. G41 lipases were stable in presence of hydrophobic organic solvents [2,5]. Oceanobacillus rekensis PT- 11 lipase was stable in presence of long-chain alcohols [4]. Lipase from Idiomarina sp. W33 and Haloarcula sp. G41 have been successfully used for biodiesel production. Protein structural studies to support organic solvent tolerant stability in halophilic lipases, are however not available. The present study addresses this gap.
Halophilic Marinobacter sp. EMB5 showing extracellular lipolytic activity was reported from our lab previously which showed significant levels of lipase secretion [8]. The present work describes the purification and characterization of the naturally aggregated lipase of this strain that showed remarkable organic solvent stability. The organic solvent stability of the lipase was further investigated by secondary and tertiary structure studies in presence of a stabilizing as well as destabilizing organic solvent.
Materials and Methods
Materials
Sephacryl S200 and p-nitrophenyl esters were a product of the Sigma Chemical Company (St. Louis, MO, USA). Polyacrylamide and bis-acrylamide were procured from GE Healthcare Life Sciences (Bangalore, India). Sodium dodecyl sulphate (SDS) and Bovine serum albumin (BSA) was purchased from Sisco Research Laboratories Pvt. Ltd. (Mumbai, India). Other media components were purchased from Hi-Media Laboratories (Mumbai, India). All other chemicals used were of analytical grade. All the solvents used in the present study were treated with molecular sieve (4 Å) prior to use.
Methods
Strain isolation, identification and lipase production: Marinobacter sp. EMB5 was isolated fromthe sea coast of Kozhikode (Kerala, 11°25′N 75°77′E) by salt enrichment. It was found to be related to Marinobacter sp. by the 16S rDNA sequence analysis which was submitted to GenBank, NCBI, USA with accession number FJ525429 [8]. Lipase was produced in a medium containing (g/l): peptone 5.0; yeast extract 5.0; NaCl 50.0; gum arabic 1.0 and olive oil 5.0 ml/l. The pH of the medium was maintained at 9.0. The production medium was seeded with 4% (v/v) inoculum raised overnight in nutrient broth with 5% (w/v) NaCl (A660~1.0) and incubated at 30°C and 160 rpm for 48 h. Cell-free supernatant (CFS), containing the extracellular lipase was obtained by pelleting down the cells by centrifugation of the culture broth at 8,000 rpm for 15 min.
Lipase assay: Lipase was assayed following the method of Kilcawley et al. [9] using p-nitrophenyl octanoate as the substrate. Briefly, 1.8 ml of Tris-HCl (50 mM, pH 8.5) containing 0.5% (v/v) Triton X-100 and 200 μL of diluted enzyme was pre-incubated for 10 min following which 20 μL of 50 mM p-nitrophenyl octanoate (PNPO) dissolved in acetonitrile was added to the reaction mixture. PNPO hydrolysis was carried out for 20 minutes at 50°C. The p-nitrophenol released by enzyme action was measured at 400 nm. One unit (U) of lipase activity was defined as the amount of enzyme releasing 1 nmol of p-nitrophenol equivalent per minute from the ester under standard assay conditions.
Lipase purification and characterization: The crude lipase was subjected to ultrafiltration using 10 MWCO and retentate, so obtained was subjected to size-exclusion chromatography by loading onto a Sephacryl S200 column (1.5 cm × 50 cm) equilibrated with Tris-HCl buffer (50 mM, pH 8.5, containing 2% (w/v) NaCl) and eluted at a flow rate of 24.0 ml/h. Fractions of 2 mL each were collected using the fraction collector (BioRad Model 2110, California, USA). Each fraction was assayed for lipase activity. The active fractions were pooled and characterized.
Disaggregation of EMB5 lipase: The Marinobacter sp. EMB5 lipase aggregates were treated with 70% 2-propanol, which was included in the SEC elution buffer as per the method of Dunhaupt et al. [10] (Table 2).
Protein determination, electrophoresis and zymogram analyses: Protein content was determined using the Bio-Rad Bradford Assay Kit (BioRad, California, USA) with bovine serum albumin as the standard. In order to determine the molecular mass of the lipase, Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was carried out according to the method of Laemmli [11]. A 12.5% SDS-PAGE gel was used on a BioRad Model Mini Protean gel electrophoresis unit (BioRad, California, United States). Silver staining was used to visualize protein bands on gel. Relative molecular mass (Mr) was estimated by comparison with molecular mass standard (14-97.4 kDa) (Bangalore Genie, Bangalore, India). Zymography was done by detection on lipolytic bands on the SDS-PAGE gel using tributyrin as substrate [12].
Characterization of EMB5 Lipase
Effect of temperature and pH on lipase activity and stability
The optimum temperature for enzyme activity was determined by assaying at various temperatures ranging between 30°C and 60°C. Thermal stability was determined by pre-incubating the enzyme at various temperatures (50°C-70°C) for different time periods and then measuring the residual lipase activity. Effect of pH on lipase activity was determined by assaying in buffers (50 mM) of varying pH viz. sodium acetate (pH 4.0-5.0), sodium phosphate (pH 6.0-7.0), Tris-HCl (pH 8.0-10.0) and Borax-NaOH buffer (pH 10.0-12.0). The effect of pH on stability was investigated by preincubating the lipase at various pH (ranging from 4.0 to 12.0) for 12 h at 25°C. The residual activity was determined under standard assay conditions.
Determination of kinetic constants
Km and Vmax values of the purified lipase were determined by assaying at different p-nitrophenyl octanoate (PNPO) concentrations. Kinetic constants were calculated using the Line weaver-Burk plot.
Effect of metal ions and additives
Lipase activity was studied in presence of 1 and 5 mM of metal salts viz. Na+, K+, Mg2+, Ca2+, Zn2+, Cu2+, Hg2+ and additives phenylmethylsulfonyl fluoride (PMSF), p-chloromercuribenzoic acid (PCMB), Dithiothreitol (DTT), β-mercaptoethanol and Ethylene diamine tetra acetate (EDTA).
Surfactant stability
The effect of detergents, Sodium dodecyl sulfate (SDS), Cetyl trimethylammonium bromide (CTAB), Tween 80, Triton X-100 and Brij 35 at 1% (v/v) concentration, was examined by incubating the enzyme with these detergents at 30°C for 5 h. The residual activity was then measured by standard assay procedure.
Substrate specificity
Substrate specificity of lipase was checked against 50 mM of p-nitrophenyl esters of acetate (C2), butyrate (C4), caprylate (C8), caprate (C10), laurate (C12), myristate (C14), palmitate (C16) and stearate (C18) under the standard assay conditions.
Organic solvent stability
One milliliter of lipase was mixed with equal volume of organic solvents (ethanol, isopropanol, butanol, dichloromethane, benzene, toluene, cyclohexane, hexane and decane) in screw capped tubes. The mixture was incubated at 30°C with constant shaking at 200 rpm for 24 h. Samples were withdrawn from aqueous phase and the residual enzyme activity estimated. Enzyme incubated without solvent was treated as control and its activity was taken as 100%.
Far UV CD spectroscopy
In order to study the effect of solvents on the secondary structure, the far UV CD spectrum of the control, cyclohexane and 2-propanol treated enzyme samples was recorded on a JASCO J-815 spectrometer (Jasco Corporation, Tokyo, Japan) using a cell with a path length of 0.1 cm and a scanning rate of 50 nm min−1 with a 2 nm bandwidth over the wavelength range 200-250 nm for far-UV with five scans averaged for each spectrum using a protein concentration of 0.5 mg/ml in the respective buffers. All CD spectra were corrected for buffer contributions.
Fluorescence spectroscopy
The effect of cyclohexane and 2-propanol on the protein tertiary structure was studied from the fluorescence spectra recorded at 25°C on a Perkin Elmer LS50 spectrofluorimeter (PerkinElmer, MA, USA). The fluorescence emission spectra were recorded from 300 to 400 nm upon excitation at 295 nm in presence of cyclohexane and 2-propanol. These organic solvents were chosen based on the organic solvent stability of the lipase, which was highest in cyclohexane and least in 2-propanol. The excitation and emission slit widths were kept at 2.5 nm and 5 nm, respectively. All fluorescence spectra were normalized and corrected for buffer contributions. All the experiments were done in triplicate and the variation was within ± 5%.
Results and Discussion
Halophiles have been perceived as a potential source of industrially useful enzymes endowed with exceptional stabilities in high salt, alkaline pH and organic solvents [13,14]. The isolation of the halophile Marinobacter sp. EMB5 strain, from Kozhikode sea water, India, has been previously reported by us [8]. The isolate was Gram-negative rod and grew well at high salt concentrations (1-20%, w/v) and pH range of 6.0-9.0. Identification of the isolate by 16S rDNA analysis related it to genus Marinobacter. The sequence has been submitted in GenBank, NCBI, USA with accession number FJ525429.
The isolate was found to be a potent lipase producer. Lipase production was growth dependent. The Cell-free Supernatant (CFS) collected at 48 h was used for all purification and characterization studies. The crude was quite stable in cyclohexane and therefore an interesting system to understand the structural basis of this phenomenon in halophiles.
Lipase purification
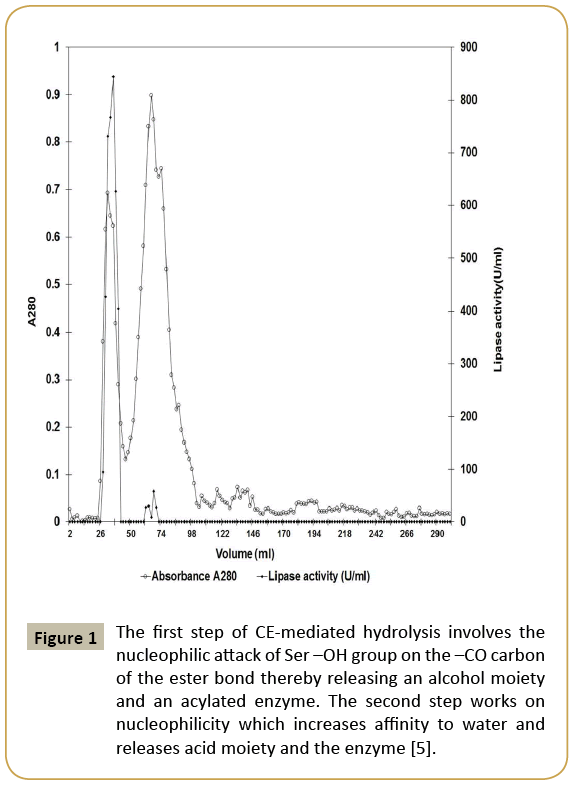
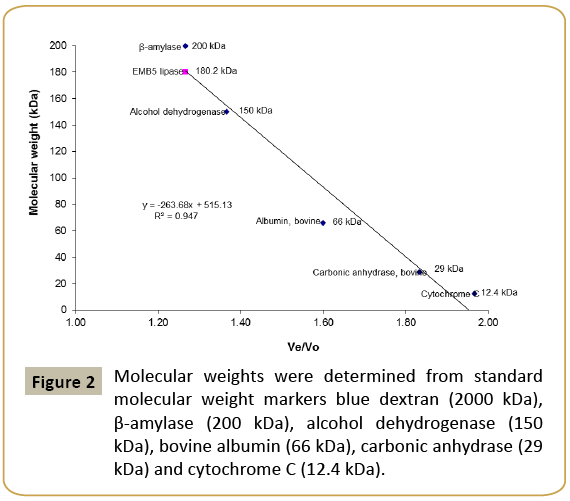
Since very few lipases are purified from halophiles, Marinobacter sp. EMB5 lipase was first attempted to be purified by conventional gel chromatography using Sephacryl S200 (Table 1). The purification chromatogram is shown in Figure 1. The lipase activity was eluted close to the void volume, indicating possibility of high molecular weight lipase aggregates. Aggregation in non-halophilic lipases is well known and has been widely studied [15,16]. In the extreme halophile Haloarcula marismortui, an esterase with a tendency to aggregate in presence of high salt concentration has been previously described by Rao et al. [17]. Although aggregation is less expected in halophiles because the presence of high salt concentration in the cytoplasm may prevent aggregation of proteins. The Marinobacter sp. EMB5 lipase also shows the same phenomenon wherein despite presence of 2% (w/v) NaCl in the CFS, lipase aggregation is observed. These lipase aggregates are soluble, active and have a high molecular mass of 180 kDa (Figure 2).
Figure 1: The first step of CE-mediated hydrolysis involves the nucleophilic attack of Ser –OH group on the –CO carbon of the ester bond thereby releasing an alcohol moiety and an acylated enzyme. The second step works on nucleophilicity which increases affinity to water and releases acid moiety and the enzyme [5].
| Purification step | Total activity | Total protein | Yield | Specific activity | Fold purification |
|---|---|---|---|---|---|
| (U) | (mg) | (%) | (U/mg) | ||
| Crude | 115874 | 200 | 100 | 579.37 | 1.0 |
| Ultrafiltration (10000 MWCO) |
88418 | 20 | 76.3 | 4420.9 | 7.6 |
| Sephacryl S200 | 6525 | 0.4 | 5.6 | 16312.5 | 28.2 |
Table 1: Purification of Marinobacter sp. EMB5 lipase.
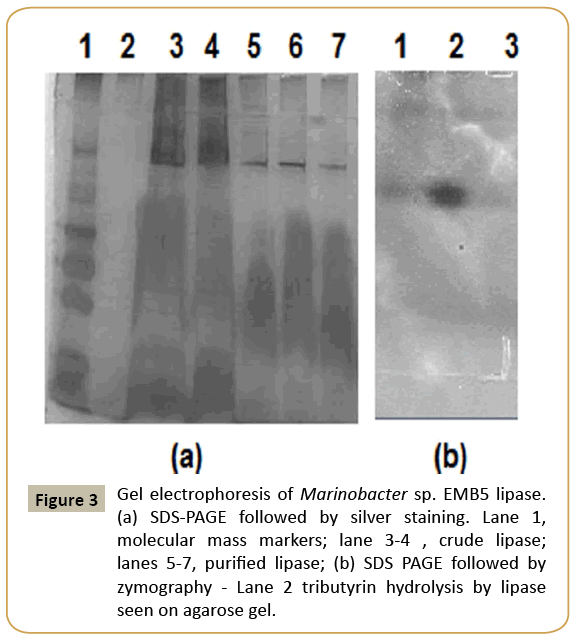
Previous studies on disaggregation of other lipases have shown that certain additives such as 2-propanol, sodium cholate, lauryldimethylamine-N-Oxide (LDAO) and SDS successfully dissociate them into monomeric forms (Table 2). The Marinobacter sp. EMB5 lipase aggregates were disaggregated by the inclusion of 70% 2-propanol in the elution buffer of Sepharcyl S200 gel chromatography. The lipase peak fractions obtained from SEC were pooled, concentrated against sucrose and subjected to SDS-PAGE. A single protein band confirmed the homogeneous purification of lipase with a molecular mass of approx. 82 kDa on the SDS PAGE gel (Figure 3a). Similar molecular weight lipases have been reported from non-halophiles, Vibrio and Burkholderia species and strains of Pseudomonas fluorescens (https://www. ncbi.nlm.nih.gov/protein/?term=LIPASE). The zymography also revealed the presence of hydrolysis zone in the tributyrin agar plate confirmed it to be a lipase (Figure 3b).
Figure 3: Gel electrophoresis of Marinobacter sp. EMB5 lipase. (a) SDS-PAGE followed by silver staining. Lane 1, molecular mass markers; lane 3-4 , crude lipase; lanes 5-7, purified lipase; (b) SDS PAGE followed by zymography - Lane 2 tributyrin hydrolysis by lipase seen on agarose gel.
| Source organism for lipase | Additive and its concentration | Molecular Mass of monomer (kDa) | Reference |
|---|---|---|---|
| Pseudomonas cepacia | 70% (v/v) 2-propanol | 33 | [9] |
| Bacillus thermocatenulatus | 1% (w/v) Sodium cholate | 40 | [24] |
| Bacillus thermocatenulatus | 5% (v/v) 2-propanol and 2 mM LDAO | 40 | [14] |
| Thermosyntrophalipolytica | 1% (w/v) SDS | 56 | [25] |
| Marinobactersp.EMB5 | 70% 2-propanol | 10 | Present study |
*Molecular mass of monomer, as determined by gel filtration chromatography.
Table 2: Effect of various additives on lipase aggregation.
Thus, our study confirms the aggregation phenomenon in Marinobacter lipase despite the presence of 2% NaCl. The ultrafiltration (10,000 MWCO) followed by Sephacryl S200 gel exclusion chromatography in presence of 70% (v/v) 2-propanol led to disaggregation of lipase aggregates. Various combinations of chromatographic procedures have been employed for purification of halophilic lipases in the past. Two steps led to 2.5 fold purification with 15% yield in halophilic lipase from Oceanobacillus strain PT-11 [4]. Ammonium sulphate precipitation using 75% (w/v) ammonium sulphate followed by DEAE- Sepharose anion exchange and Sephacryl S-200 gel filtration chromatography resulted in 7.4 purification fold with 24.6% yield in case of Idiomarina sp. W33, extracellular lipase [2]. Thus, our strategy to purify monomer (disaggregated form) by incorporating 2-propanol and to purify aggregated form by omitting 2-propanol worked well.
Characterization of purified lipase
The purified aggregated lipase showed 16312.5 U/mg specific activity while the disaggregated showed 208.28 U/mg. Considering that (i) lipase is more active in aggregated form, as observed in case of other lipases too [18] and (ii) that from the application point of view, the purified active lipase was characterized.
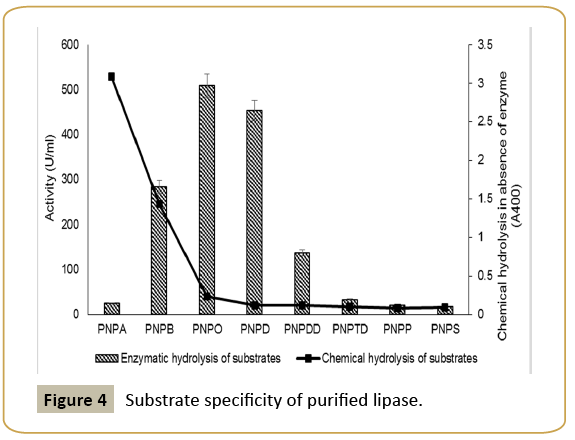
Enzymatic properties: The enzymatic characteristics of Marinobacter have not been investigated in details before. The enzyme exhibited broad substrate specificity with maximum activity towards C8 and C10 substrates (Figure 4). Similar substrate specificity has been reported in case of Salicola sp. IC10 [19] and LipBL from Marinobacter lipolyticus lipa [6] lipases. It indicates that Marinobacter sp. lipase in the present case has better affinity for medium chain length fatty acids.
The Km and Vmax of the lipase towards p-nitrophenyl octanoate (PNPO) were 1.43 μM and 1.43 μmoles/ml/min respectively. Among other enzymatic properties, the EMB5 lipase had temperature optima at 50°C and pH optima at 9.0. The lipase was stable in the pH range 8.0-10.0 up to 12 h. Thus the Marinobacter lipase was alkaline in nature. These are in agreement with other halophilic lipases. Lipase LipBL from Marinobacter lipolyticus was stable over the pH range of 7.0-10.0 [6] but that from Oceanobacillus was stable only in a narrow range between pH 8.0-9.0 [4].
As a very novel feature, the Marinobacter lipase was significantly heat stable as compared to other reported halophilic lipases. It had a half-life of 90 minutes at 60°C and 30 min at 70°C. This property makes it quite attractive for applications at elevated temperatures. Among the lipases which have their temperature optimum at 50°C, Aspergillus terreus lipase is stable at 60°C for more than 12 h [20].
Effect of additives and metal ions: The effect of metal ions and additives (oxidizing, reducing and chelating agents) was tested at 1 and 5 mM in 0.1 M Tris-HCl buffer at pH 8.5. Lipase was activated by Ca2+ ions and was inhibited by the presence of K+, Cu2+, Hg2+ and Ba2+. The activation in presence of Ca2+ ions was even more pronounced at 5 mM concentration. Lipases in general are known to be activated by Ca2+ [13]. The lipase was less responsive to PMSF, indicating the non-involvement of serine, unlike other lipases. However, PCMB caused a decrease in activity, indicating involvement of cysteine residue.
Organic solvent and surfactant stability: Solvent stability is a generic feature of all halophilic proteins [13,21]. This is attributed to the fact that they work at high salt surroundings which is akin to low water medium. Lipase in the present study was stable in 50% (v/v) concentration of cyclohexane (Table 3). Polar solvents are known to strip off essential water of activity from active site universally causing loss in activity, so was in the present case. Among the halophilic lipases, solvent stability has been reported from Haloarcula, Idiomarina, Alkalibacillus salilacus [2,3,5].
| Solvent | Log P value | Relative activity in 50% (v/v) after 24 h (%) |
|---|---|---|
| Control | - | 100 |
| Ethanol | -0.235 | 29 |
| Isopropanol | 0.05 | 23 |
| Butanol | 0.8 | 44 |
| Cyclohexane | 3.2 | 116 |
| Hexane | 3.9 | 77 |
| Decane | 6.0 | 16 |
Table 3: Stability of Marinobacter sp. lipase in organic solvents.
Surfactant stability: The lipase was found to be extremely stable in presence of CTAB, Triton X100, Tween 80 and Brij 35 except anionic SDS which is a highly desirable property for lipase from the viewpoint of detergent applications (Table 4). The halophilic lipases have hydrophobic amino acid residues in the lid that covers the lipase active site. The organic solvent molecules interact with these residues and are likely to maintain the enzyme in the open conformation. This may contribute mainly to their stability in presence of organic solvents and surfactant stability [22]. This is the first study which reports both these properties for Marinobacter lipase, making it quite attractive for applications.
| Surfactant | Relative activity after 5 h at 1% (v/v) (%) |
|---|---|
| Control | 100 |
| CTAB | 190 |
| Triton X100 | 131 |
| Tween 80 | 153 |
| Brij 35 | 141 |
| SDS | 64 |
Table 4: Stability of Marinobacter sp. lipase in surfactants.
Structural Studies
Far UV CD spectroscopy
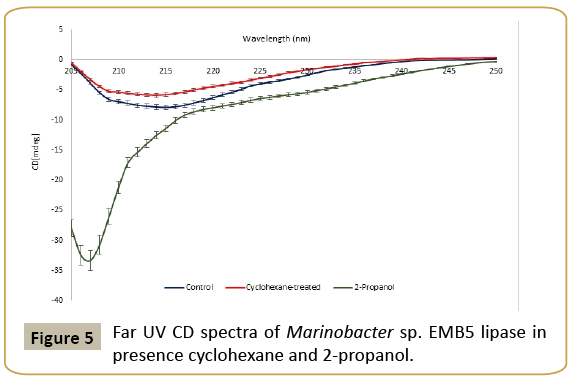
In order to understand solvent stability, the CD spectra of purified lipase were recorded in presence and absence of cyclohexane at 25°C and pH 8.5 (Figure 5). Marinobacter lipase was extremely stable in presence of cyclohexane up to 24 h. The results show changes in the peak position of negative ellipticity between 222 and 225 nm. This region corresponds to the spectral profile of α-helix [23]. In presence of cyclohexane, there was only a minor decrease in negative ellipticity as compared to the control. Thus, the CD spectra have given us an insight into the secondary structure stability in presence of an organic solvent in which the lipase displays enormous stability. Despite a very high concentration of the organic solvent and a prolonged exposure to solvent, there were no distinct changes in the secondary structure.
Fluorescence spectroscopy
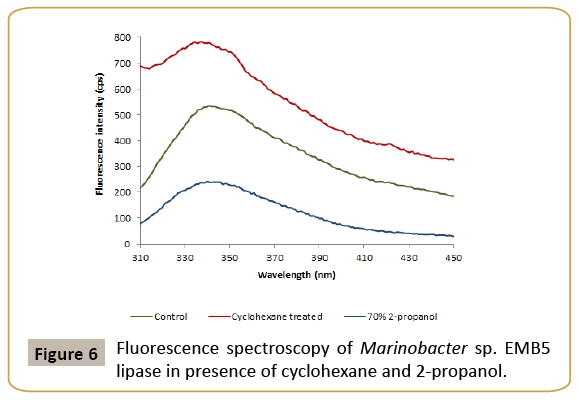
The effect of cyclohexane on the tryptophan fluorescence spectrum of lipase was studied by exciting the samples at 295 nm (Figure 6). The solvated tryptophan residues in aqueous solutions have fluorescence maxima at 350 nm. Under conditions that promote protein folding, the tryptophan residues are firmly embedded in the non-polar protein core causing a characteristic blue shift of its fluorescence spectrum. Denaturing solvents, on the other hand cause an increase in fluorescence intensity with a characteristic red shift of tryptophan fluorescence in the 328 to 348 nm wavelength range. Incubation of EMB5 lipase with cyclohexane caused a red shift from 344.5 nm to 346.5 nm. This suggested a slight increase of the accessibility of these residues to the solvent, i.e., increase of the protein molecule hydrodynamic volume, evidently fluorescence quenching in presence of cyclohexane. Similar phenomenon has been previously reported by us in case of halophilic protease in presence of n-heptane and n-decane [7]. Presence of a biphasic solvent system of water and cyclohexane causes conspicuous changes in the immediate microenvironment of the lipase. However, due to the presence of salt we expect these changes to have minimal effect on the interactions that stabilize the protein structure. In fact, decreasing fluorescence intensity is suggestive of tryptophan residues still buried in the EMB5 lipase hydrophobic core which is part of the enzyme active site. This explains the stability of the halophilic enzyme even in presence of a high cyclohexane concentration. Most probably the interactions responsible for the functional tertiary structure are more or less intact and there is very little if any loss of structure. The results are in agreement with secondary structure data to suggest that the structural changes whatsoever are minimal and do not result in compromising enzyme function.
Conclusions
Halophilic isolate Marinobacter sp. EMB5 produced a lipase with interesting surfactant and organic solvent stability. The lipase is prone to aggregation. It was purified 28.2 fold by sizeexclusion chromatography on Sephacryl S200 column. The active form of lipase occurred in aggregated form. The active form lipase was stable in a wide range of organic solvents at 50% (v/v) concentration even up to 24 h. This property was well correlated to its retaining its secondary and tertiary features. It was haloalkaline in nature, having an optimum pH at 9.0 and salt 2% (w/v), respectively. It was stable across a wide pH range of 5.0 to 10.0 and temperature up to 60°C. Thus, the Marinobacter sp. EMB5 lipase emerges as a novel halo-alkaline lipase which could be successfully purified by Sephacryl S200.
Acknowledgements
The financial support by the Department of Biotechnology (Government of India) No. BT/PR4935/PID/6/655/2012 is gratefully acknowledged. Author RH is grateful to the Indian Council of Medical Research (ICMR), India, for Research Fellowship No. 3/1/3/JRF-2010/ MPD-47(36165). The technical assistance provided by the staff in the Central Instrumentation Facility of the Department of Chemistry, Indian Institute of Technology Delhi is gratefully acknowledged.
Conflict of Interest
The authors RH and SKK declare that they have no conflict of interest.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Bisht D, Yadav SK, Darmwal NS (2013) An oxidant and organic solvent tolerant alkaline lipase by P. aeruginosa mutant: downstream processing and biochemical characterization. Braz J Microbiol 44: 1305-1314.
- Li X, Qian P, Wu SG, Yu HY (2014) Characterization of an organic solvent-tolerant lipase from Idiomarina sp. W33 and its application for biodiesel production using Jatropha oil. Extremophiles 18: 171-178.
- Samaei-Nouroozi A, Rezaei S, Khoshnevis N, Doosti M, Hajihoseini R, et al. (2015) Medium-based optimization of an organic solvent-tolerant extracellular lipase from the isolated halophilic Alkalibacillus salilacus. Extremophiles 19: 933-947.
- Jiewei T, Zuchao L, Peng Q, Wang L, Yongqiang T (2014) Purification and characterization of a cold-adapted lipase from Oceanobacillus strain PT-11. PLoS One 9: e101343.
- Li X, Yu HY (2014) Characterization of an organic solvent-tolerant lipase from Haloarcula sp. G41 and its application for biodiesel production. Folia Microbiol 59: 455-463.
- Perez D, Martin S, Fernandez-Lorente G, Filice M, Guisan JM, et al. (2011) A novel halophilic lipase, LipBL, showing high efficiency in the production of eicosapentaenoic acid (EPA). PLoS One 6: e23325.
- Sinha R, Khare SK (2014) Protective role of salt in catalysis and maintaining structure of halophilic proteins against denaturation. Front Microbiol 5: 165.
- Kumar S, Karan R, Kapoor S, Singh SP, Khare SK (2012) Screening and isolation of halophilic bacteria producing industrially important enzymes. Braz J Microbiol 43: 1595-1603.
- Kilcawley K, Wilkinson M, Fox P (2002) Determination of key enzyme activities in commercial peptidase and lipase preparations from microbial or animal sources. Enzyme Microb Technol 31:310-320.
- Dunhaupt A, Lang S, Wagner F (1992) Pseudomonas cepacia lipase: studies on aggregation, purification and on the cleavage of olive oil. Biotechnol Lett 14: 953-958.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685.
- Oh BC, Kim HK, Lee JK, Kang SC, Oh TK (1999) Staphylococcus haemolyticus lipase: biochemical properties, substrate specificity and gene cloning. FEMS Microbiol Lett 179: 385-392.
- Moreno ML, Perez D, Garcia MT, Mellado E (2013) Halophilic bacteria as a source of novel hydrolytic enzymes. Life 3: 38-51.
- Schreck SD,Grunden AM (2014) Biotechnological applications of halophilic lipases and thioesterases. Appl Microbiol Biotechnol 98: 1011-1021.
- Schlieben NH, Niefind K, Schomburg D (2004) Expression, purification and aggregation studies of His-tagged thermoalkalophilic lipase from Bacillus thermocatenulatus. Protein Expr Purif 34: 103-110.
- Velu N, Divakar K, Nandhinidevi G, Gautam P (2012) Lipase from Aeromonas caviae AU04: Isolation, purification and protein aggregation. Biocatal Agric Biotechnol 1: 45-50.
- Rao L, Zhao X, Pan F, Li Y, Xue Y, et al. (2009) Solution Behavior and Activity of a Halophilic Esterase under High Salt Concentration. PLoS One 4: e6980.
- Timucin E, Sezerman OU (2013) The Conserved Lid Tryptophan, W211, Potentiates Thermostability and Thermoactivity in Bacterial Thermoalkalophilic Lipases. PLoS One 8: e85186.
- Moreno ML, Garcia MT, Ventosa A, Mellado E (2009) Characterization of Salicola sp. IC10, a lipase- and protease-producing extreme halophile. FEMS Microbiol Ecol 68: 59-71.
- Littlechild J, Novak H, James P, Sayer C (2013) Mechanisms of thermal stability adopted by thermophilic proteins and their use in white biotechnology. In: Thermophilic Microbes in Environmental and Industrial Biotechnology, Biotechnology of Thermophiles, Second Edition. Springer Science & Business Media.
- Gupta A, Khare SK (2009) Enzymes from solvent-tolerant microbes: Useful biocatalysts for non-aqueous enzymology. Crit Rev Biotechnol 29: 44-54.
- Rua L, Diaz-Maurino T, Fernandez VM, Otero C, Ballesteros A (1993) Purification and characterization of two distinct lipases from Candida cylindracea. Biochim Biophys Acta 1156: 181-189.
- Kelly SM, Jess TJ, Price NC (2005) How to study proteins by circular dichroism. Biochim Biophys Acta 1751: 119-139.
- Rua ML, Schmidt-Dannert C, Wahl S, Sprauer A, Schmid RD (1997) Thermoalkalophilic lipase of Bacillus thermocatenulatus: large-scale production, purification and properties: aggregation behaviour and its effect on activity. J Biotechnol 56: 89-102.
- Salameh MDA, Wiegel J (2010) Effects of detergents on activity, thermostability and aggregation of two alkalithermophilic lipases from Thermosyntropha lipolytica. Open Biochem J 4: 22-28.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences