Protein Hox Serves as the Transcriptional Guarantee for the Asset of Cell Fate
Sadaf Javed* and Farukh Azeem
Department of Bioinformatics and Biotechnology, Government Collage University Faisalabad, Faisalabad, Pakistan
- *Corresponding Author:
- Sadaf Javed
Department of Bioinformatics and Biotechnology,
Government Collage University Faisalabad,
Faisalabad,
Pakistan,
E-mail:sadafsadafjaved@gmail.com
Received Date: April 19, 2021;Accepted Date: May 03, 2021;Published Date: May 10, 2021
Citation: Javed S,AzeemF (2021) Protein Hox Serves as the Transcriptional Guarantee for the Asset of Cell Fate.J Mol Biol BiotechVol.6 No.3:02.
Abstract
HOX genes that encode evolutionary number of transcription factors that regulate how an organism's phenotype is arranged by its genetic composition. While HOX genes are considered to control the differentiation of Stem Cells (SC) and HOX genes are poorly controlled in cancer, there is a lack of understanding of mechanisms through which inhibition of HOX genes in SCs induces production of the cancer. The aim of this study was to examine the function of HOX genes in stem cell differentiation, specifically in embryo, infant tissue, and pluripotent stem cells and to explore the responsibilities of dysfunctional HOX genes in SCs for colorectal cancer, acute myeloid (leukaemia) production. Hox gene consists of four different clusters (Hoxa, Hoxc, Hoxb and Hoxd) and composed of 13 paralog pairs that present on four different chromosomes. The entire task of the Hox gene goes beyond the boundary of this study, but the ultimate purpose is to decide an organism's front and back body plans. 39 Hox gene components organized into HoxA, HoxC, HoxB, and HoxD were present in the mouse. The development of the phylogenetic tree is commonly used to organize new sequences of Hox into their classes. Another automatic tool is to assign Hox sequence according to the maximum sequence similarity by using BLAST technique. This profile is a matrix transcription factors that enables a score to be assigned to a series on the basis that it resembles the profile. This actually offers considerable public materials for future computer study of Hox genes, specifically in groups whereby limited taxon samples have demonstrated limitations to conclusions reached for the whole population for example, sponges without Hox, ParaHox genes. Hox pred will become an art method for the automated detection and classification of Hox genes from complete genome analysis.
Keywords
HOX gene family; Homeodomain; Transcription factors; Human phenotypes; Transition; Locus growth regulator
Introduction
The homeobox comprising the genes is a significant family of evolutionary and conserved regulators that regulate the embryonic growth of the planned animal body. In Drosophila’s hox genes were first identified where homosexuality or morphological transition of one part of the body into another were found to be responsible. Eight genes of Drosophila Hox have all contained a retained 180 base pair fragment of DNA (bp). This 'homeobox' covers the homeodomain, a DNA binder protein DNA helix style with a motif of 60 amino acids, which contains the larger and lesser ribs of its target location for the DNA recognition (Lufkin, 2000). Many other genes that possessed a homeodomain, however, were recently described, which differed and are thus not part of the same Hox family. The homeodomain amino acid sequences and subsequent protein domains have permitted the classification of homeo domain containing homeoprotein into more than 20 different families (Hox, Hmx, Dlx, PBC, Hmx, and PBC) with 2-10 mammalian members each family. This is significantly higher for the special Hox family that comprises 39 participants of mammals [1,2]
Enhancer and repressor components control Hox genes. While Hox genes are prominently regulating corporeal morphology, current research studies investigate the expression of Hox genes in the nervous system of vertebrate organisms. Studies have found that the Hox gene plays a special function in the neuronal distinction and recognition of axons in response to environmental indicators. XHoxc10 was stated to be upregulated at the location of the recruitment of distinguished and unidentifiable cells. In the multiple biological processes Hox gene perform an important role. The lack of the Hox gene will actually cause significant deficiencies. For instance, Hoxa3's disruption results in thymus failure. The loss of Hoxa1 or hoxd11, Hoxc11 may also lead to the destruction of met nephric renal function [3-8].
In its entirety, the homeodomain is known a motif and a standard profile. The relationship between this motif and Exd or PBX class of Hox Cofactors (known collectively as PBC) has been demonstrated through biochemical, structural and functions analysis. The 2nd domain, called UbdA (UA), is primarily present in AbdA and Ultrabithorax central Hox proteins (Ubx). This Para log domain was newly introduced Exd selection in repression of the seen to be essential Gene that promotes limb. Domain three (TD), the YPWM motif also similar in series (TDWM), specific prologue. The TD Motive preserves the W, which gives the PBC category proteins close interaction and correlates the HX motif sequence with certain Hox Proteins. Today evolutionary and creation biologists continue to be fascinated by the remarkable deep preservation of the genetic organisation, its role and fundamental effect on the evolution of metaso an organism plans [9-12].
The research focuses on many possible forms of Hox biology, including regulating the Hox gene, identifying downstream objectives, and revealing new protein roles. Hox genes have also been linked to a variety of human diseases which supports an intensified need to consider their possible role in the onset and disease progression. Future science needs to start using genetic methods to integrate greater diversity of species to gather a better understanding of the impacts of the Hox cluster. Currently though, most Hox analyses rely on a single species with developments limited to similarities with previously reported records. In a single sample, a better more impartial strategy is to investigate many organisms and then use explicit techniques to evaluate alternate hypothesis. Finally, we studied the HOX gene literature and its associated therapeutics, which may help us understand how SC-specific therapy is developed, aiming at abnormal HOX gene regulation that helps cancer [13-17].
Materials and Methods
Identification of HOX GENE
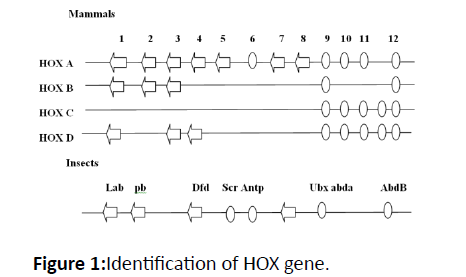
NCBI has been used to derive protein and DNA sequences of Hox genes from Protein blasting was carried out using a human gene HOX protein sequence. Alignment with other species was carried out. Variants with The online resources and SMART were used in order to exclude any sequences which have not preserved motifs. The longest open read frames were chosen and their E value tested attentively. The sequences processed through these databases gave the domain conserved sequences. The NCBI website has provided information such as accession numbers, chromosome location, gene locus, amino acid numbers, and the length of open reading frames in Figure 1.
Domains and motifs identification
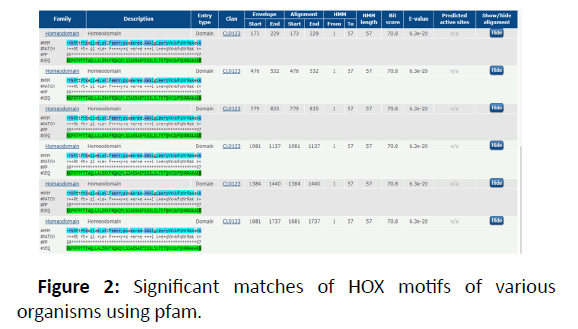
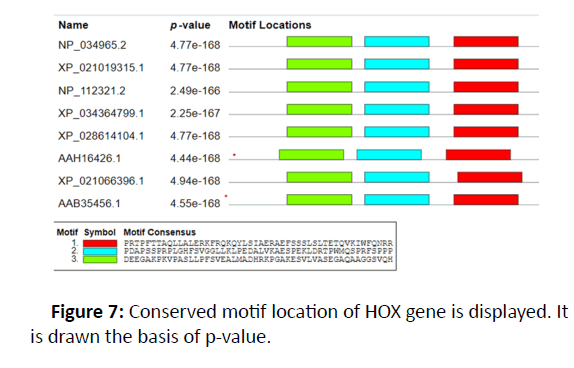
In order to study preserved protein patterns and domains at the Online Pfam NCBI Database, the HOX protein sequence was supplied as a query sequence and all target sequences have been downloadedand smart. This step has been taken with sequences in which no conserved domains and motif have been retained which are important for the normal performance of HOX protein. The number of motif was picked as 8 and the pattern distribution was chosen as independent, meaning that the motif could be repeated. A gene structure display server was used to display exons and introns in the HOX gene in Figure 2 [4,5].
Phylogenetic analysis of HOX gene family
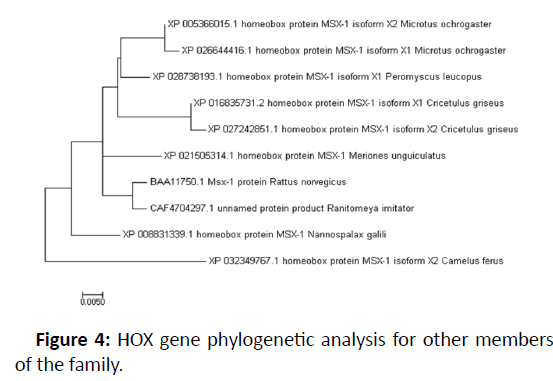
It was made using programme Mega 7.0 for the phylogenetic tree. The bioinformatics tool MEGA7 with its default parameters was used to align every sequence for the reporting of conserved domains in the HOX gene members with multiple alignment and pair-wise alignment. The nearest family relatives in the HOX gene were retrieved from the Gene bank database series. MEGA7 has been used to organize sequences and on the sequence alignment bases a perfect un-rooted evolutionary tree was developed. In all amino acids in each sequence 4 per cent divergence had been detected and this value was denoted as a bar at the base of the tree. The HOX gene family was also categorised of its evolutionary similarities after that. The evolutionary family relationships were carried out using the Time Tree Server online in Figures 3 and 4 [6,7].
Analysis of gene structure (introns and exons)
The HOX gene structure was analysed using an online instrument known as the gene structure display, which was used to determine the location of introns and exons and obtained a three-dimensional protein structure in Figure 5 [18-24].
Results and Discussion
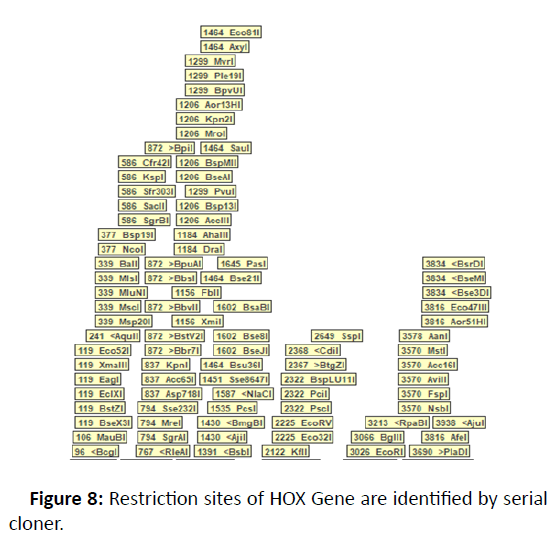
There were 15 distinct HOX genes found in Homo sapiens with various methods and databases, with additional detail, including RNA, DNA and protein sequences, provided in the table of evidence in Figures 6-8. The evolutionary connection reveals that different genes are identical. On chromosome number 7, the gene location was identified. Four exons and one domain are identified in Table 1.
| Proposed names | HOX |
|---|---|
| Gene locus | HOXA10 |
| Protein accession # | NP_061824.3 |
| RNA accession# | NM_018951.4 |
| Exons | 3 |
| Chr # | 2 |
| ORF length | 1230 |
| Amino acid length | 410 |
| Start of genomic location | 7p15.2 |
Table 1: Nomenclature suggested and significant characteristics of HOX genes.
Conclusion
Hox proteins direct tissue morphology involving animal embryo and act as both transcription activators and repressors which likely have an impact on the representation of many responsive genes. Hox genes are the targets for automobile and inter regulation, regulated by the evolved regulatory proteins of DNA in the clusters of Hox gene. The objective of this analysis is to identify different germ line mutation Phenotypes in those 10 Hox genes that affect a human phenotype, with particular focus on the genotypic and phonotypical differences across allelic disorders. With the increasing usage in the future of clinical entire led to the expansion and genome sequence, we expect that other Hox genetic mutations will certainly affect different humanphenotypes. The homozygous mouse lack of functionality phenotypes for 29 Hox genes with no established human disorders are also checked. This study will allow physicians to detect and treat patients with identified Hox gene disease and help them determine the potential for new mutations in individuals with mouse knockout trials based phenotypes.
References
<- Acampora D, D'esposito M, Faiella A, Pannese M, Migliaccio E, et al. (1989) The human HOX gene family. Nucleic Acids Res 17(24):10385-10402.
- Arnold CP, Migueles Lozano A, Mann FG, Lange JJ, Seidel C, et al. (2021) Hox genes regulate asexual reproductive behavior and tissue segmentation in adult animals.
- Bhatlekar S, Fields JZ, Boman BM. (2018) Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int 2018:3569493.
- Cillo C. (1994) HOX genes in human cancers. Invasion Metastasis 14(1): 38-49.
- Francis JC, Gardiner JR, Renaud Y, Chauhan R, Weinstein Y, et al. (2021) HOX genes promote cell proliferation and are potential therapeutic targets in adrenocortical tumours. 124(4):805-816.
- Gehring WJ. (2007) The homeobox as a key for understanding the principles of the genetic control of development. HOX Gene Expression.
- Hahn JM, McFarland KL, Combs KA, Anness M, Supp DM. (2021) Analysis of HOX gene expression and the effects of HOXA9 overexpression in fibroblasts derived from keloid lesions and normal skin. Wound Repair Regen.
- Hrycaj SM, Wellik DM. (2016) Hox genes and evolution. F1000Res.
- Krumlauf R. (1994) Hox genes in vertebrate development. 78(2):191-201.
- Boiteux EL, Court F, Guichet PO, Vaurs‐Barrière C, Vaillant I, et al. (2021) Widespread overexpression from the four DNA hypermethylated HOX clusters in aggressive glioma is associated with H3K27me3 depletion and alternative promoter usage. Mol Oncol 2021
- Li B, Huang Q, Wei GH. (2019) The role of HOX transcription factors in cancer predisposition and progression. Cancers. 11(4):528
- Lufkin T. (2005) Hox Genes: Embryonic Development.
- Mallo M, Alonso CR. (2013) The regulation of Hox gene expression during animal development. Development 140(19):3951-3963
- Morgan R, Plowright L, Harrington KJ, Michael A, Pandha HS. (2010) Targeting HOX and PBX transcription factors in ovarian cancer. BMC Cancer 10:89
- Murray J, Preston E, Crawford JP, Rumley JD, Amom P, et al. (2021) The anterior Hox gene ceh-13 and elt-1/GATA activate the posterior Hox genes nob-1 and php-3 to specify posterior lineages in the C. elegans embryo.
- Nakashoji A, Hayashida T, Yamaguchi S, Kawai Y, Kikuchi M, et al. (2021) Comprehensive analysis of the homeobox family genes in breast cancer demonstrates their similar roles in cancer and development. Breast Cancer Res Treat 186(2):353-361.
- Postika N, Schedl P, Georgiev P, Kyrchanova O. (2021) Mapping of functional elements of the Fab-6 boundary involved in the regulation of the Abd-B hox gene in Drosophila melanogaster. Scientific Reports 11(1):4156.
- Procino A, Cillo C. (2013) The HOX genes network in metabolic diseases. Cell Biol Int 37(11):1145-1148.
- Quinonez SC, Innis JW. (2014) Human HOX gene disorders. Mol Genet Metab 111(1):4-15.
- Ruddle FH, Bartels JL, Bentley KL, Kappen C, Murtha MT, et al. (1994) Evolution of Hox genes. Annual Review of Genetics 28:423-442.
- Tang D, Zhang Z, Zboril E, Wetzel MD, Xu X, et al. (2021) Pontin Functions as A Transcriptional Co-activator for Retinoic Acid-induced HOX Gene Expression.
- Tanzer A,Amemiya CT, Kim CB, Stadler PF. (2005) Evolution of microRNAs located within Hox gene clusters. J Exp Zool B Mol Dev Evol 304(1):75-85.
- Taylor HS. (2000) The role of HOX genes in human implantation. Hum Reprod Update 6(1):75-79.
- Thomas-Chollier M. (2016) Computational analysis of transcriptional regulation in metazoans. Ecole normale supérieure-ENS PARIS
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences