ISSN : 2573-4466
Insights in Enzyme Research
Properties Of Two Novel Esterases Identified From Culture Supernatant Of Penicillium Purpurogenum Grown On Sugar Beet Pulp
Gabriela Oleas1, Eduardo Callegari2, Romina Sepulveda1 and Jaime Eyzaguirre1*
1Facultad de Ciencias Biologicas, Universidad Andres Bello, Santiago, Chile
2BRIN-USDSSOM Proteomics Facility, University of South Dakota, Vermillion,SD, USA
- *Corresponding Author:
- Dr. Jaime Eyzaguirre
Facultad de Ciencias Biologicas, Universidad
Andres Bello, Republica 217, Santiago, Chile.
Tel: +56226618070
E-mail: jeyzaguirre@unab.cl
Received Date: November 28, 2016; Accepted Date: December 09, 2016; Published Date: December 12, 2016
Citation: Oleas G, Callegari E, Sepulveda R, et al. Properties of Two Novel Esterases Identified from Culture Supernatant of Penicillium purpurogenum Grown on Sugar Beet Pulp. Insights Enzyme Res. 2016, 1:1. doi: 10.21767/2573-4466.100004
Abstract
Background: The filamentous fungus Penicillium purpurogenum grows on a variety of natural carbon sources, such as sugar beet pulp, and secretes to the medium a large number of enzymes that degrade the carbohydrate components of lignocellulose. Sugar beet pulp is rich in pectin, and the purpose of this work is to identify novel esterases produced by the fungus, which may participate in pectin degradation.
Methods and findings: Partially purified culture supernatants of the fungus grown on sugar beet pulp were subjected to mass spectrometry analysis. Peptides thus identified, which may be part of potential esterases were probed against the proteins deduced from the fungal genome sequence. The cDNAs of two putative esterases identified were expressed in Pichia pastoris and their properties studied. One of these enzymes, named FAET, is a feruloyl esterase, while the other, PE, is classified as a pectin methyl esterase.
Conclusions: These findings add to our knowledge of the enzymology of pectin degradation by Penicillium purpurogenum, and define properties of two novel esterases acting on de-esterification of pectin. Their availability may be useful as tools for the study of pectin structure and degradation.
Keywords
Esterases; Penicillium purpurogenum; Heterologous expression; Pichia pastoris; Sugar beet pulp
Abbreviations
pNP: p-nitrophenyl; DTNB: 5,5′-dithiobis(2-nitrobenzoic acid); MUA: Methylumbelliferyl acetate; PMSF: Phenyl methyl sulfonyl fluoride; RG I: Rhamnogalacturonan I.
Introduction
Pectin is one of the polysaccharides present in lignocellulose, and it is found mainly in the primary cell wall of plants [1]. Several components can be identified in pectin structure: 1) Homogalacturonan (a polymer of D-galacturonic acid residues bound by α-1, 4 linkages, which may be methylated and acetylated). 2) RG I (formed by a backbone of alternating α-D-galacturonic acids and L-rhamnoses, linked to arabinan and galactan side-chains and showing acetyl and methyl esterification), and 3) rhamnogalacturonan II (formed by short chains of galacturonic acid residues substituted by complex sidechains of a variety of monosaccharide’s) [1]. The biodegradation of pectin is a complex process, in which a number of different glycanases, esterases and lyases participate [2].
In our laboratory, we have used for a number of years the filamentous fungus Penicillium purpurogenum as a model for the study of the enzymology of lignocellulose biodegradation. This fungus grows on different lignocellulose-containing substances, and secretes to the medium a large number of enzymes, which degrade the lignocellulose polysaccharides [3,4]. Among the carbon sources used is sugar beet pulp. This product is composed of 20% cellulose and 50% pectin [5]. In this work, we have aimed at the finding of potential esterases, which may participate in pectin degradation. As a result, two esterases named FAET and PE have been identified, heterologously expressed in Pichia pastoris and characterized.
Methods
Microbial strains utilized
P. purpurogenum strain ATTC MYA-38 was utilized as the source of enzymes. For cloning purposes, E. coli DH5 α or TOP10F were used, and heterologous expression was performed in Pichia pastoris GS115, supplied in the Easy Select Pichia Expression Kit (Invitrogen, CA, USA).
Liquid cultures of P. purpurogenum
P. purpurogenum was grown on Mandels medium as described previously [6] using sugar beet pulp or glucose at 1% as carbon sources. Liquid cultures on sugar beet pulp (12 L) were incubated for 7 days at 28°C in a shaker at 200 RPM. After filtration on cheesecloth and centrifugation, the clear supernatant was concentrated using a Minitan (Millipore, U.S.A) ultrafiltration apparatus (10 KDa cutoff membrane) and Amicon (Millipore) ultrafiltration Centricons, to a final volume of about 20 mL.
Partial purification of esterases
In order to obtain a crude separation of esterases, the concentrated supernatant was subjected to chromatography. A pseudo-affinity chromatography was performed using Cibacron blue Sepharose: 50 mL of resin were equilibrated in 50 mM citrate buffer pH 4 and 15 mL of the concentrated supernatant were added. The column was washed with 3 volumes of the buffer and then 3 volumes of a gradient (0 to 1 M NaCl) in the buffer were used to elute proteins.
Zymograms
Polyacrylamide gels were prepared according to Laemmli [7]. Samples (concentrated chromatography column eluates) contained 25 to 100 μg protein in 60 mM Tris HCl buffer pH 6.8, 25% glycerol and 0.1% Bromophenol Blue. The samples were loaded directly on the gel and the electrophoresis was performed at 4°C for 3 hours at 80 V. The gels were then washed 3 times with 50 mM acetate buffer pH 5 and were then placed in 100 mL of the same buffer containing 1 mg MUA (dissolved in absolute ethanol). The appearance of fluorescence bands was photographed under UV light. The gels were then washed with buffer and stained with Coomassie Brilliant Blue. The bands showing enzyme activity were cut and subjected to mass spectrometry analysis.
Mass spectrometry and bioinformatics analysis
Mass spectrometry was performed by tandem LC/MS/MS as described by Dong et al. [8]. The resulting data were analyzed by MASCOT v. 2.4 using a protein database derived from the P. purpurogenum genome (unpublished). Peptides showing a score >18 were considered. The amino acid sequence of each protein found was analyzed by CDD SEARCH (https://www.ncbi. nlm.nih.gov/Structure/cdd/cdd.shtml) in order to establish which proteins contained domains present in esterase families. In addition, each protein identified as potential esterase was subjected to BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) to search for homologies with other possible esterases. Alignment with homologous sequences was performed using CLUSTAL Omega (https://www.ebi.ac.uk/ Tools/msa/clustalo/). Gene and cDNA sequences were identified using the program Augustus (https://bioinf.uni-greifswald.de/ augustus/). Molecular weights and isoelectric points were estimated with EXPASY (https://www.expasy.org/); the presence of a signal peptide was determined with SignalP (https://www. cbs.dtu.dk/services/SignalP/). dbCAN (https://csbl.bmb.uga.edu/ dbCAN/) was used to assign the protein sequences found to the CAZY database families (https://www.cazy.org/). Presence of Oand N-glycosylation was analyzed using NetOGlic and NetNglic (https://www.cbs.dtu.dk/services/NetOGlyc/ and https://www. cbs.dtu.dk/services/NetNGlyc/), respectively.
DNA preparation
P. purpurogenum was grown on 100 mL of 1% glucose for 5 days. The mycelium obtained was separated by centrifugation, frozen in liquid nitrogen and pulverized with a mortar and pestle. DNA was extracted by means of the Genomic DNA Purification kit (Fermentas). The manufacturer’s instructions were followed in all procedures using kits.
PCR and overlap extension PCR
PCR was performed with either Taq polymerase (Thermo Scientific) for routine assays and High Fidelity PCR Enzyme mix (Thermo Scientific) for cloning and sequencing. The conditions were as follows: initial denaturation at 95°C for 5 min and 30 cycles of denaturation at 95°C for 30 sec, annealing at 59°C for 30 sec, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were gel purified.
Introns were eliminated using “overlap extension PCR” as described by Topakas et al. [9]. The PCR conditions used were the same as those indicated above. Table 1 lists the primers used for amplification of whole cDNAs and Table 2 those used for overlap extension PCR.
| Gene | Primers |
|---|---|
| faet | FAETini: ATGCGGTTTTCGCTG (sense) FAETfin: TTAAACACACTTCCAAGCATTC (antisense) |
| pe | PEini: ATGCATTTGACAACGCTCCTTTCAGC (sense) PEfin: CTAATAACTCGAATCAATCCAATCACCATA (antisense) |
Table 1: Primers used for amplification of esterase cDNAs.
| Gene | Primers |
|---|---|
| faet | FAETini: ATGCGGTTTTCGCTG (sense) FAETfin: TTAAACACACTTCCAAGCATTC (antisense) FAETOR: ATGTCAGAGTATTGAATACAGCCACCGAGGCCT (antisense) FAETOF: CCTCGGTGGCTGTATTCAATACTCTGACATGGTCT (sense) |
| pe | PEini: ATGCATTTGACAACGCTCCTTTCAGC (sense) PEfin: CTAATAACTCGAATCAATCCAATCACCATA (antisense) PEOF1: AGGGTGCTCAGGCTGTTGCACTCGTCGGCAACGC (sense) PEOR1:TATTGGCGTTGCCGACGAGTGCAACAGCCTGAGCA (antisense) PEOF2: CAACTGCCTGATCGAAGGAGCCGTGGACTACAT (sense) PEOR2: CGAAGATGTAGTCCACGGCTCCTTCGATCAGGCAG (antisense) |
Table 2: Primers utilized for “Primer Extension PCR.”
Cloning and expression in Pichia pastoris
Cloning of the PCR products was performed in pGEM-T-Easy (Promega). Competent E. coli DH5 α cell were transformed with the resulting plasmids. Transformed cells were plated in LB agar supplemented with 1 μL 50 mg/mL Ampicillin and 1 μL 20 mg/ mL X-Gal. Plasmids from white colonies were extracted using the Plasmid DNA Mini Kit II (Omega Bio Tek) and used as templates for PCR with primers including restriction enzymes recognition sites (Table 3) for directional cloning. The PCR products obtained were cloned in pPICZB and the resulting plasmids were transformed into competent E. coli TOP10F. Transformed cells were grown in LB plates containing 100 μg/mL Zeocin. The presence of inserts in the resulting clones was checked by PCR, and they were sequenced in both strands using the Sanger method (Macrogen Inc., Seoul, Korea). The DNA sequences of the genes coding for the two enzymes have been entered in GenBank under the following numbers: faet: KP313782; pe: KP313784.
| Gene | Sense | Antisense |
|---|---|---|
| faet | P1.43F: GCGGCCGCATGCGGTTTTCGCTG | P1.43R: GAATGCTTGGAAGTGTGTTTAATCTAGACGC |
| pe | P2.61F: CGAGAATTCATGCATTTGACAACGCTCCTT | P2.61R: GTGATTGGATTGATTCGAGTTATTAGTCTAGACGC |
The recognition sequences for restriction enzymes are underlined
Primer (sense) for FAET includes a NotI recognition site.
Primers (sense) for PE contain an EcoRI recognition site.
All antisense primers include a restriction site for XbaI.
Table 3: Primers with restriction enzyme recognition sites.
The plasmids of pe were linearized by means of SacI, while those of faet, by PmeI, and transformed into Pichia pastoris by electroporation. Transformed clones were selected in YPD (Yeast extract Peptone Dextrose medium) agar plates supplemented with 100 μg/mL Zeocin. DNA was extracted from the clones to confirm by PCR the presence of the insert. The positive clones were grown in 20 mL BMGY (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base with ammonium sulfate without amino acids, 0.02% biotin and 1% glycerol) for 2 to 3 days (200 RPM, 28°C) to obtain sufficient biomass. The cultures were centrifuged and re-suspended in 20 mL of BMMY medium (glycerol is replaced by methanol) to induce enzyme production, incubating for up to 5 days. Aliquots were removed to assay for enzyme activity. Larger scale enzyme production was performed (500 mL volume) with the clones of interest.
Pectin extraction from sugar beet pulp
The procedure described by Ralet et al. [10] was used. The extraction was performed with water in order to preserve possible methyl and acetyl esters present in the pectin.
Enzyme Assays
Qualitative assay of esterase activity
For this purpose, MUA (a substrate utilized by many esterases) was utilized. One mg of MUA (Sigma) was dissolved in 1 mL ethanol and 9 mL 50 mM acetate buffer pH 5 were added. Assays were performed in micro plate wells, using 200 μL of substrate and 20 to 50 μL of enzyme preparation. The formation of fluorescent methyl umbelliferone was followed under UV light.
Assays with pNP derivatives
The following substrates were utilized: pNP-acetate, pNPbutyrate, pNP-decanoate, pNP-dodecanoate, pNP-myristate, pNP-palmitate (all from Sigma) and pNP-ferulate (Carbosynth). 1 mL stock solution of each compound (100 mM) was prepared in dimethyl sulfoxide. They were used to prepare dilutions (1 to 10 mM) in 100 mM sodium phosphate buffer pH 5.8. Tests were performed in microplates, using between 1 to 20 μg proteins in a total volume of 200 μL. Reactions were incubated at 28°C for 15 or 30 min and absorbance at 405 nm were measured immediately.
Qualitative assay for pectin methyl esterase with Ruthenium Red (Merck)
The technique described by Chakiath et al. [11] was employed. Various pectins (all from Sigma) were used as substrates: citrus pectin 3%, 55-70% and 85% methyl esterified and apple pectin 70-75% methyl esterified. Agar plates containing the substrates were added 5-10 μL enzyme and then incubated at 37°C from 30 min to 2 hours. The plates were washed and covered with 3 mL 0.5% Ruthenium Red. After 1 min the plates were washed. The presence of red spots indicates positive activity. Ruthenium red binds preferably to the anionic polygalacturonic acid rather than to the methyl-esterified pectin [11].
Detection of tannases activity with rhodanine
The assay was performed according to Sharma et al. [12] using methyl gallate (Sigma) as substrate. 480 μL 0.01 M substrate in 50 mM citrate pH 5 were added 20 μg protein from culture supernatants. After 30 min at 30°C, 300 μL rhodanine (0.667 g in 100 mL methanol) were added, and the mixture incubated for 5 min at 30°C before addition of 200 μL 0.5 M KOH. Absorbance at 520 nm was measured after an additional incubation (30° for 5 min).
Liberation of acetic acid from pectin
Reaction mixtures containing 160 μL 50 mM phosphate buffer pH 7, 20 μL 10% sugar beet pulp pectin and 20 μL enzymes were incubated for 18 h at 28°C. Acetate liberation was detected using the Acetic Acid (K-ACETRM) kit from Megazyme. As positive control, an alkaline hydrolysate of the pectin was used; the method proposed by Juturu et al. [13] for acetylated xylan was followed.
Quantitative assay for pectin methyl esterase activity
The procedure described by Grsic-Rausch and Rausch [14-19] was followed, using 50 mM phosphate buffer pH 7.4. Increase of absorbance (NADH formation) was followed at 340 nm after 15-30 min incubation.
Protease activity assay
As substrate, 100 mM azo-casein (Sigma) in 50 mM phosphate buffer pH 6 was used. 250 μL azo-casein were mixed with 150 μL enzyme and incubated for 30 min at 37°C. Reaction was stopped with 1.2 mL 1% trichloroacetic acid. The mixture was centrifuged, the supernatant was added 1.4 mL 1 M NaOH and absorbance was measured at 440 nm.
Inhibition by PMSF
A 100 mM solution of PMSF was prepared in absolute ethanol. Enzyme activity was measured using either pNP-acetate as substrate or the pectin methyl esterase assay, with a final PMSF concentration of 2 mM.
Structure modeling
The amino acid sequences of the identified proteins were used to construct homology models using Schrodinger’s Prime suite program (https://www.schrodinger.com/Prime/) after elimination of the signal peptides (detected by SignalP). The templates used are indicated for each enzyme in the Results section.
Results
Identification of possible esterases in sugar beet pulp culture supernatant
Culture supernatants were subjected to partial purification by chromatography in Cibacron-Blue-Sepharose. Activity was found (using the qualitative MUA assay) in both the washing and the eluate.
Samples of the active fractions obtained in the chromatography were analyzed for MUA activity using zymograms. The bands showing activity were cut and subjected to mass spectrometry analysis. As a result, the genes of two potential esterases (named FAET and PE) were identified. FAET was found in the washing (Mascot score of 19), and PE in the gradient of the chromatography (Mascot score of 88). Both DNA sequences contain introns which were eliminated by Overlap Extension PCR. The resulting cDNA’s were sequenced and the results coincide with the sequences annotated in the genome.
Heterologous expression in Pichia Pastoris
The cDNAs were linked to pPICZB, the resulting plasmids were linearized and were transformed into Pichia pastoris. Two clones of each protein were selected and subjected to enzyme induction by methanol. Activity of the supernatants was assayed with MUA; only the clones of FAET showed activity. When assayed for pectin methyl esterase activity (with methyl esterified pectin using the Ruthenium Red method) only the PE clones were active. No activity for both assays was shown by the negative control (P. pastoris transformed with the empty vector).
Properties of FAET
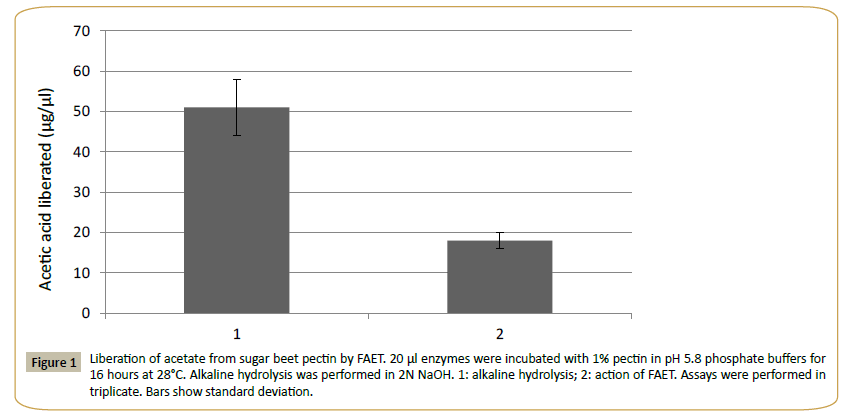
The theoretical molecular mass of FAET was estimated at 56.381 kDa and the pI was estimated at 5.2. The protein presented a signal peptide and five N-glycosylation sites and 3 O-glycosylation sites were identified. The enzyme was active on MUA, pNPacetate (KM=2.3 mM) and pNP-ferulate (KM=1.4 mM). The assay with pNP-ferulate was used for optimum pH and the assay with pNP-acetate for optimal temperature determinations. The pH was tested in the range of pH 2.0 to 12.0 (using citrate, Tris- HCl and carbonate buffers), and the effect of temperature in the range from 20°C to 80°C at pH 5.8. The results gave a pH optimum near 8 and an optimal temperature of 38°C. According to the CDD database, FAET belongs to the tannase superfamily. However, FAET did not possess tannase activity when assayed with rhodanine. The enzyme was capable of liberating acetic acid from sugar beet pulp pectin (Figure 1). The control culture of P. pastoris lacking the FAET insert showed none of the activities presented by FAET.
Figure 1: Liberation of acetate from sugar beet pectin by FAET. 20 μl enzymes were incubated with 1% pectin in pH 5.8 phosphate buffers for 16 hours at 28°C. Alkaline hydrolysis was performed in 2N NaOH. 1: alkaline hydrolysis; 2: action of FAET. Assays were performed in triplicate. Bars show standard deviation.
Properties of PE
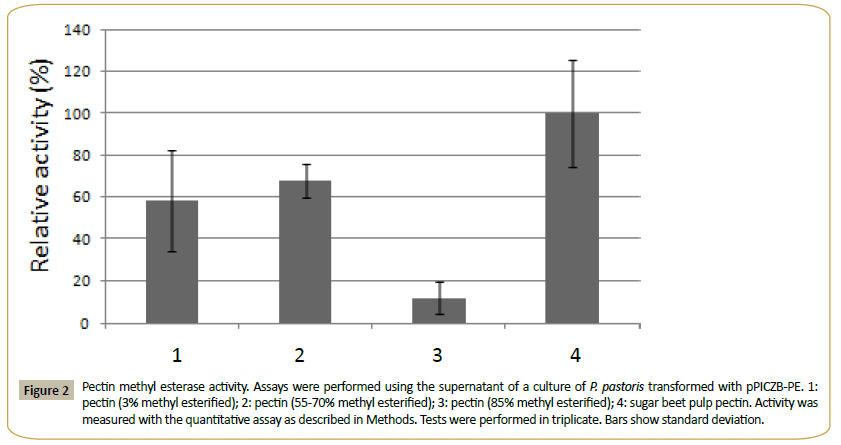
This enzyme had calculated molecular mass of 32.199 kDa and an estimated pI of 5.4. One N-glycosylation site and 11 O-glycosylation sites were identified, and the enzyme showed a signal peptide. PE presented no activity over MUA and the pNP derivatives of acetate, ferulate, butyrate, myristate, decanoate, laureate and palmitate. It was active in the methyl esterase qualitative test with Ruthenium Red, and using the quantitative assay (see Methods) on citrus pectin with different degrees of methyl esterification and on sugar beet pulp pectin (Figure 2). The quantitative method was used to determine the pH optimum (in the range from pH 3.0 to 9.0) and the temperature optimum (range of 25°-65°C) of PE. Using 3% methyl esterified pectin as substrate, a pH optimum of 5.0 and an optimal temperature of 30°C were obtained for the enzyme. The supernatant of the culture of P. pastoris transformed with the plasmid without insert showed none of these activities.
Figure 2: Pectin methyl esterase activity. Assays were performed using the supernatant of a culture of P. pastoris transformed with pPICZB-PE. 1:pectin (3% methyl esterified); 2: pectin (55-70% methyl esterified); 3: pectin (85% methyl esterified); 4: sugar beet pulp pectin. Activity was measured with the quantitative assay as described in Methods. Tests were performed in triplicate. Bars show standard deviation.
Discussion
Faet
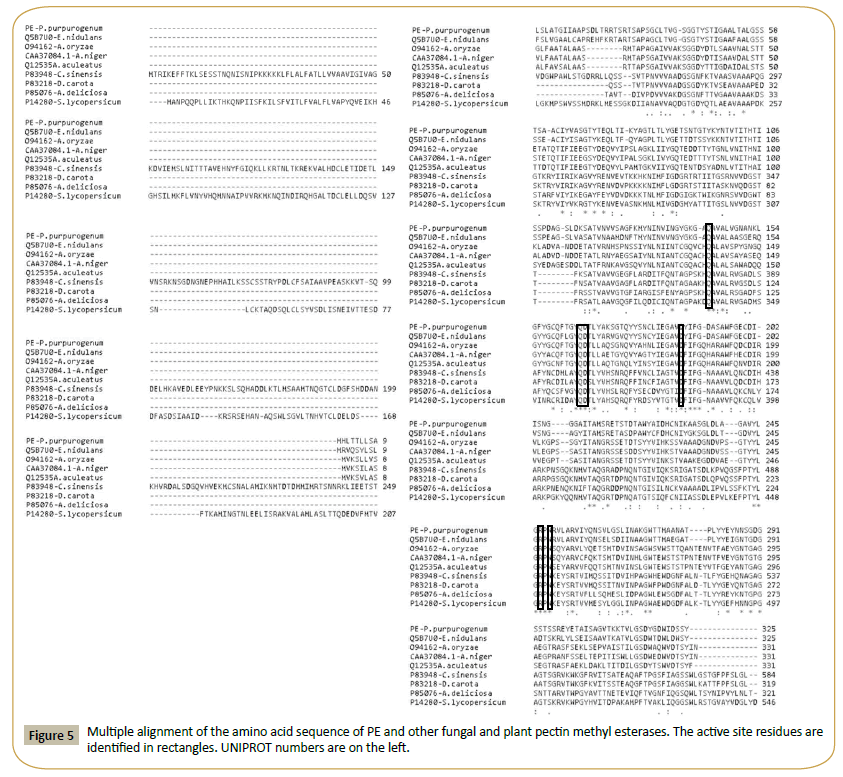
A BLASTP analysis of the amino acid sequence of FAET shows 6 characterized feruloyl esterases with sequence identities ranging from 72% down to 42% (Table 4). All these enzymes belong to the tannase superfamily and possess a tannase domain (Pfam 07519) but lack tannase activity. Only one enzyme is known with both feruloyl esterase and tannase activities; it was isolated from a metagenomic library obtained from a soil sample [20], and it shows only 26% identity with FAET. The sequences of these enzymes are aligned in Figure 3, including, for comparison, a tannase from Aspergillus oryzae [21]. The fungal feruloyl esterases and the tannase from A. oryzae present the nucleophilic elbow (GXSGX) characteristic of the α/β hydrolases, but it is absent in the enzyme from metagenomic origin. All the sequences possess the motif CS-D-HC, which has been found to be essential for the A. oryzae feruloyl esterase activity [22]. dbCAN assigned FAET to CAZy family CE1; however, FAET shows a very low identity with enzymes from this family: only 22% identity is found with a feruloyl esterase from Aspergillus nidulans (GenBank: EAA62427). More recently, new classifications of feruloyl esterases have been introduced based on phylogenetic analysis [23,24]; due to its high identity (72%) to the Aspergillus oryzae AoFaeB, (UNIPROT Q2UP89), FAET can be classified in family SF1.
Figure 3: Multiple alignment of the amino acid sequence of FAET with feruloyl esterases and tannases. The catalytic triad is highlighted in rectangles and the nucleophilic elbow is underlined. The residues forming the motif CS-D-HC are marked with vertical arrows. UNIPROT identification numbers for each sequence are shown on the left.
| Organism (UNIPROT code N°) | References | % Identity with FAET | Optimum pH | Optimal temperature (°C) |
|---|---|---|---|---|
| FAET | This work | 100% | 8 | 38 |
| Aspergillus oryzae (Q2UP89) | [15] | 72% | 6 | - |
| Aspergillus oryzae (Q2UMX6) | [15] | 57% | 6 | - |
| Aspergillus niger (Q8WZI8) | [16] | 50% | 6 | - |
| Aspergillus nidulans (Q5BCF8) | [17] | 54% | 7 | 45 |
| Talaromyces stipitatus (Q70Y21) | [18] | 53% | 6-7 | 60 |
| Fusarium oxysporum (W9NXZ3) | [19] | 42% | 6 | 65 |
Table 4: Comparison of the properties of FAET with feruloyl esterases of similar sequence.
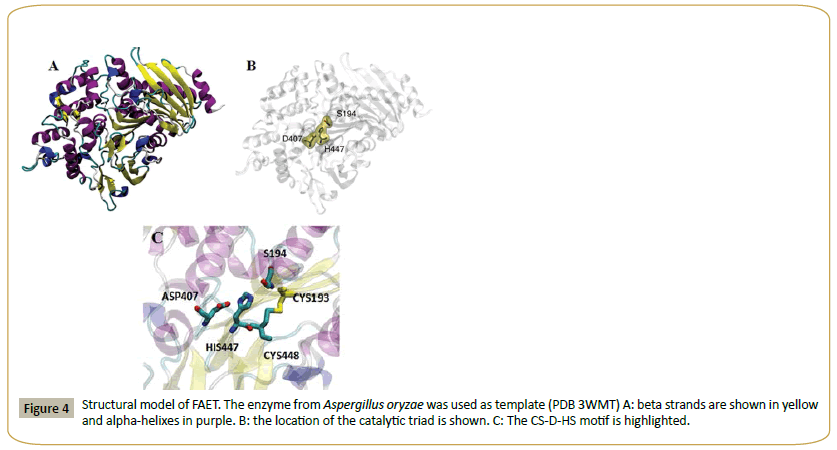
A. oryzae AoFaeB has been crystallized and its structure determined [22-29]. This structure has been used as a template to build a model for FAET (Figure 4). The structure shows the catalytic triad (S194, D407, H447). That FAET is a serine esterase has been confirmed, since the enzyme is completely inactivated by 2 mM PMSF. The enzyme lacks protease activity, since it is inactive on azo-casein. In short, FAET can be defined as a feruloyl esterase from family SF1.
Figure 4: Structural model of FAET. The enzyme from Aspergillus oryzae was used as template (PDB 3WMT) A: beta strands are shown in yellow and alpha-helixes in purple. B: the location of the catalytic triad is shown. C: The CS-D-HS motif is highlighted.
PE
By a BLASTP analysis of the sequence of PE, four characterized enzymes were found with identities ranging from 70% to 46%. A comparison of their properties is shown in Table 5. Analysis by dbCAN assigns PE to CAZy family 8, which includes only pectin methyl esterases, in agreement with CDD, which assigns it to the pectin esterase superfamily.
| Enzyme source (UNIPROT code N°) | References | % identity with PE | Optimum pH | Optimal temperature (°C) |
|---|---|---|---|---|
| Penicillium purpurogenum | This work | 100% | 5 | 30 |
| Emericella nidulans (Q5B7U0) | [25] | 70% | 8 | 30 |
| Aspergillus aculeatus (Q12535) | [26] | 44% | 4.6 | 45 |
| Aspergillus niger (GENBANK CAA37084.1) | [27] | 43% | - | - |
| Aspergillus oryzae (O94162) |
[28] | 46% | 5 | 55 |
Table 5: Comparison of the properties of PE with those of other characterized fungal pectin methyl esterases.
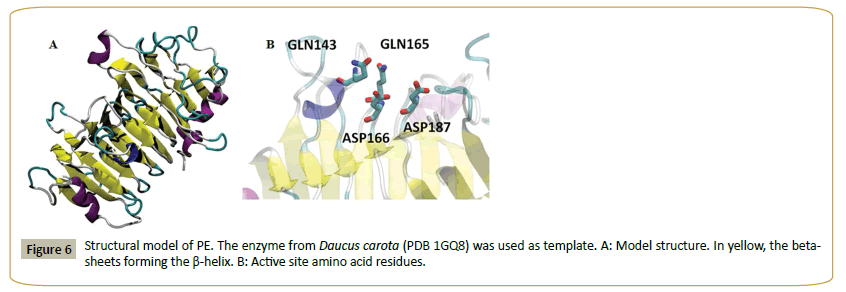
Figure 5 shows the alignment of PE and a set of fungal and plant pectin methyl esterases. Plant sequences are included, since the enzyme from carrot (Daucus carota, 31% identity to PE) [29] was used as template to model the structure of PE (Figure 6). The model shows a beta helix structure. Common active site residues were identified, despite of the low sequence identity: an asp residue (D 166) which acts as acid-base, a second asp (D 187) (nucleophile), two gln (Q 143 and 165) forming the anionic hole which stabilizes the transition state, an arg (R 247) and trp (W 249) involved in substrate binding. Both plant and fungi pectin methyl esterases seem to operate by a similar catalytic mechanism, despite different biological functions: plant enzymes participate in fruit ripening [29] while fungal enzymes are involved in the rotting of plant material. PE can thus be identified as a pectin methyl esterase.
In conclusion, two novel esterases have been identified, expressed and characterized. They likely participate in the biodegradation of the pectic component of sugar beet pulp in synergy with other hydrolases and lyases. FAET (feruloyl esterase) may participate in the liberation of ferulic and other cinnamic acid residues present in RG1. PE, a pectin methyl esterase liberates methanol from esters in homogalacturonan and RG1. These enzymes may be valuable tools for a detailed study of pectin degradation.
Funding
This work was supported by grants from FondoNacional de Ciencia y Tecnologia (FONDECYT N°1100084 and 1130180), Universidad Andrés Bello (DI-478-14/R and DI-31-12/R) and NIH grant number 2 P20 RR016479 from the INBRE Program of the National Center for Research Resources for the mass spectrometry analysis. The sponsors were not involved in any of the steps of the production of this manuscript.
Competing and Conflicting Interests
The authors declare no competing and conflicting interests.
References
- Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohyd Res 344: 1879-1900.
- Jayani RS, Saxena S, Gupta R (2005) Microbial pectinolytic enzymes: A review. Process Biochem 40: 2931-2944.
- Chavez R, Bull P, Eyzaguirre J (2006) The xylanolytic enzyme system from the genus Penicillium. J Biotechnol 123: 413-433.
- Perez-Fuentes C, Ravanal MC, Eyzaguirre J (2014) Heterologous expression of a Penicillium purpurogenum pectin lyase in Pichia pastoris and its characterization. Fungal Biol 118: 507-515.
- Saulnier L, Thibault JF(1999)Ferulic acid and di-ferulic acids as components of sugar-beet pectins and maize bran heteroxylans. J Sci Food Agr79: 396-402.
- Hidalgo M, Steiner J, Eyzaguirre J(1992)ß-Glucosidase from Penicillium purpurogenum: purification and properties. Biotechnol Appl Bioch 15: 185-191.
- Laemmli UK (1970)Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685.
- Dong G, Callegari EA, Gloeckner CJ, UeffingM, Wang H (2012)Prothymosin- alpha interacts with mutant huntingtin and suppresses its cytotoxicity in cell culture. J Biol Chem 287: 1279-1289.
- Topakas E, Moukouli M, Dimarogona M, Christakopoulos P(2012)Expression, characterization and structural modeling of a feruloyl esterase from the thermophilic fungus Myceliophthora thermophila. Appl Microbiol Biotechnol94:399-411.
- Ralet MC, Thibault JF, Valle DG (1991)Solubilization of sugar-beet pulp cell wall polysaccharides by extrusion-cooking. Lebensm-Wiss. Technol 24: 107-112.
- Chakiath C, Lyons MJ, Kozak RE, Laufer CS(2009)Thermal stabilization of Erwinia chrysanthemi pectin methyesterase A for application in a sugar beet pulp biorefinery. Appl Environ Microbiol 75: 7343-7349.
- Sharma S, Bhat TK, Dawra RK (2000)A spectrophotometric method for assay of tannase using rhodanine. Anal Biochem 279: 85-89.
- Juturu V, Aust C, Wu JC (2013)Heterologous expression and biochemical characterization of acetyl xylan esterase from Coprinopsis cinerea. World J Microbiol Biotechnol 29: 597-605.
- Grsic-Rausch S, Rausch TT(2004) A coupled spectrophotometric enzyme assay for the determination of pectin methyl esterase activity and its inhibition by proteinaceous inhibitors. Anal Biochem 333: 14-18.
- Koseki T, Hori A, Seki S, Murayama T, Shiono Y (2009) Characterization of two distinct feruloyl esterases, AoFaeB and AoFaeC, from Aspergillus oryzae. Appl. Microbiol. Biotechnol 83: 689-696.
- de Vries RP, vanKuyk PA, Kester HC, Visser J(2002) The Aspergillus niger faeB gene encodes a second feruloyl esterase involved in pectin and xylan degradation and is specifically induced in the presence of aromatic compounds. Biochem. J 363: 377-386.
- Shin HD, Chen RR (2007)A type B feruloyl esterase from Aspergillus nidulans with broad pH applicability. Appl. Microbiol. Biotechnol 73: 1323-1330.
- Garcia-Conesa MT, Crepin VF, Goldson AJ, Williamson G, Cummings NJ, et al. (2004) The feruloyl esterase system of Talaromyces stipitatus: production of three discrete feruloyl esterases, including a novel enzyme, FaeC, with broad substrate specificity. J Biotechnol 108: 227-241.
- Moukouli M, Topakas E, Christakopoulos P (2008)Cloning, characterization and functional expression of an alkalitolerant type C feruloyl esterase from Fusarium oxysporum. Appl. Microbiol. Biotechnol 79: 245-254.
- Yao J, Chen QL, Shen AX, Cao W, Liu YH(2013)A novel feruloyl esterase from a soil metagenomic library with tannase activity. J. Mol. Catal. B 95:55-61.
- Hatamoto O, Watarai T, Kikuchi M, Mizusawa K, Sekine H (1996)Cloning and sequencing of the gene encoding tannase and a structural study of the tannase subunit from Aspergillus oryzae. Gene 175: 215-221.
- Suzuki K, Hori A, Kawamoto K, Thangudu RR, Ishida T, et al.(2014)Crystal structure of a feruloyl esterase belonging to the tannase family: a disulfide bond near a catalytic triad. Proteins 82: 2857-2867.
- Benoit I, Danchin EGJ, Bleichrodt RJ, de Vries RP (2008) Biotechnological applications and potential of fungal feruloyl esterases based on prevalence, classification and biochemical diversity. Biotechnol. Lett 30: 387-396.
- Dilokpimol A, Makela MR, Aguilar-Pontes MV, Benoit-Gelber I, Hilden KS, et al. (2016) Diversity of fungal feruloyl esterases: updated phylogenetic classification, properties,and industrial applications. Biotechnol. Biofuels 9: 231.
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR (2006)Development and application of suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci 103: 11417-11422.
- Christgau S, Kofod LV, Halkier T, Andersen LN, Hockauf M, et al. (1996) Pectin methyl esterase from Aspergillus aculeatus: expression cloning in yeast and characterization of the recombinant enzyme. Biochem. J 319: 705-712.
- Khanh NQ, Ruttkowski E, Leidinger K, Albrecht H, Gottschalk M (1991) Characterization and expression of a genomic pectin methyl esterase-encoding gene in Aspergillus niger. Gene 106: 71-77.
- Kitamoto N, Okada H, Yoshino S, Ohmiya K, Tsukagoshi N (1999) Pectin methylesterase gene (pmeA) from Aspergillus oryzae KBN616: its sequence analysis and overexpression, and characterization of the gene product. Biosci. Biotechnol. Biochem 63: 120-124.
- Johansson K, El-Ahmad M, Friemann R, Jornvall H, Markovic O, et al. (2002) Crystal structure of plant pectin methylesterase. FEBS Lett 514: 243-249.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences