ISSN : 2393-8854
Global Journal of Research and Review
Plant Defense and the Counter Defense by Viruses
Department of Botany, Aligarh Muslim University, Aligarh-202002, U.P, India
Abstract
RNA silencing is a sequence specific RNA degradation process triggered by the formation of double stranded RNA that can be introduced by a virus or transgene. RNA silencing was first discovered in transgenic plants, where it was termed co-suppression or post transcriptional gene silencing (PTGS). In plants, it serves as an antiviral defense, and many plant viruses encode suppressors of silencing. RNA silencing has been a robust host defense mechanism against plant viruses is generally countered by virus-encoded silencing suppressors. This strategy now increasingly recognizes to be used by most plant viruses. We are presenting here an overview of the common features shared by some of the best studied plant viral silencing suppressors followed of the characteristics of the few reported plant viral suppressors.
Keywords
RNA silencing, PTGS, Antiviral defense, Silencing suppressors.
INTRODUCTION
Interest in gene silencing-related mechanisms stemmed from the early 1990s, when this phenomenon was first noted as a surprise observation by plant scientists during the course of plant transformation experiments, in which the introduction of a transgene into the genome led to the silencing of both the transgene and homologous endogenes. From these initial studies, plant biologists have continued to generate a wealth of information into not only gene silencing mechanisms, but also the complexity of these biological pathways as well as revealing their multilevel interactions with one another. The plant biology community has also made significant advancements in exploiting RNA silencing as a powerful tool for gene function studies and crop improvements.
RNA silencing (also called as posttranscriptional gene silencing PTGS) refers to a family of gene silencing effects by which the expression of one or more genes is down regulated or entirely suppressed by the introduction of the antisense RNA molecule. The most common and well- studied example is RNA interference, in which endogenously expressed micro RNA or exogenously derived small interfering RNA induces the degradation of complementary messenger RNA. It also plays an important role in defending plants against viruses. Enzymes detect double stranded RNA (that is, not normally found in cells) and digest it into small pieces that are unable to cause disease.
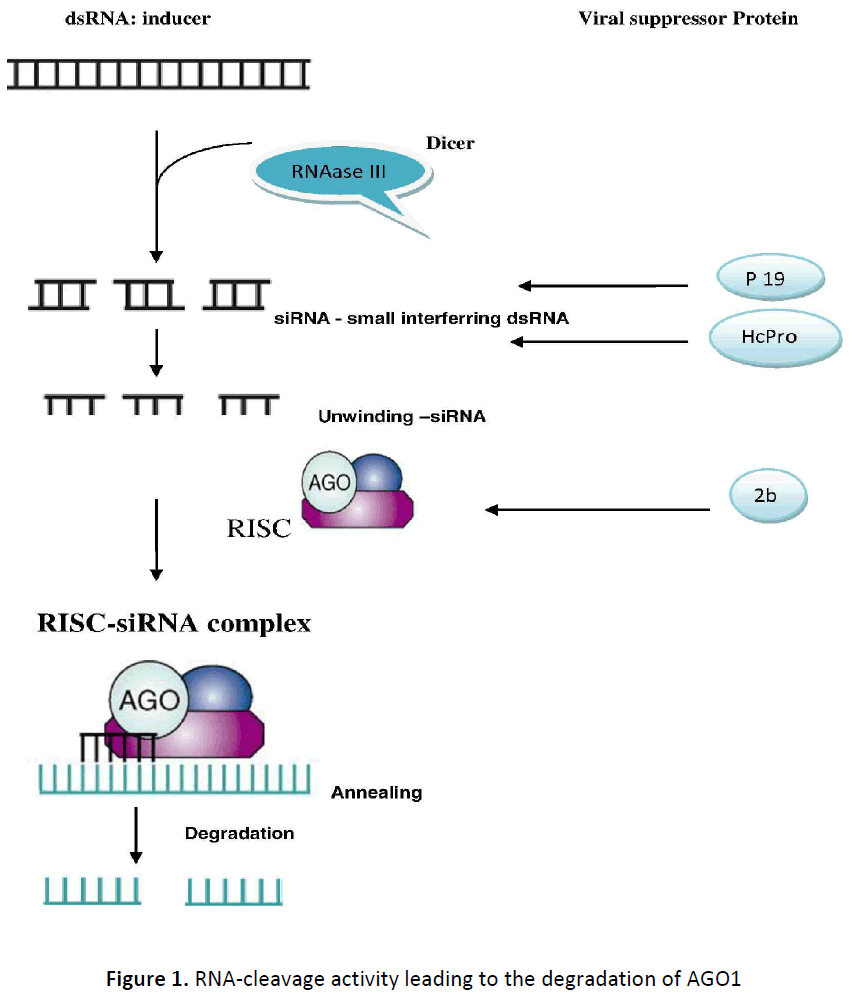
The basic RNAi process can be divided into three steps. First, a long doublestranded RNA (dsRNA) that is expressed inor introduced into- the cell (for example, as a result of the base-pairing of sense and antisense transcripts or the formation of stem–loop structures) is processed into small RNA duplexes by a ribonuclease III (RNase III) enzyme known as Dicer. Second, these duplexes are unwound, and one strand is preferentially loaded into a protein complex known as the RNA-induced silencing complex (RISC), here RISC binds to an ARGONAUTE (AGO) protein. Third, this complex effectively scans the transcriptome and finds potential target RNAs. The loaded single-stranded sRNA, called the guide strand, then directs an endonuclease that is present in the RISC (sometimes called the ‘slicer’ and now known to be an Argonaute protein to cleave messenger RNAs that contain a sequence homologous to the siRNA. The most obvious role of viral siRNAs in antiviral defense is to direct the RISC complex to viral genomic and subgenomic RNAs, thereby targeting those molecules for destruction (Figure 1 Courtesy Trends in Microbiology, Vol.16. No.5). The beauty of RNA silencing as a defense mechanism is that it couples the destruction of the dsRNA triggers of silencing with the use of the resulting siRNAs as specificity factors to target other nucleic acid molecules with complementary sequence. siRNAs can be used in several different ways as specificity factors in antiviral defense. The source for many of the dsRNA sequences in various RNA and DNA viruses is not known, although it is likely that they originate during viral replication and/or from internal pairing of long RNA molecules [5,15].
RNA Silencing is a highly complex system, composed of numerous different proteins and processes [6]. Such complexity makes this system extremely efficient for controlling not only viral infection, but also an endogenous RNA expression during plant development and growth. Nevertheless, this complexity might also make the plants vulnerable to many viruses that have developed ways of battling this defense machinery, by encoding suppressor proteins capable of interfering with various steps of RNA interference (RNA i) silencing pathway.
Salient features of RNAi
• Double stranded RNA rather than singlestranded antisense RNA is the interfering agent.
• Silencing can be introduced in different developmental stages.
• High degree of specific gene silencing with less effort.
• Avoids problems with abnormalities caused by a knocked out gene in early stages (which could mask desired observations).
• Silencing effects passed through generations.
Applications of RNA silencing
The ability to manipulate RNA silencing has a wide variety of practical applications of biotechnology ranging from molecular biology to gene therapy in animals.
• RNA interference employing short dsRNA oligonucleotides will permit to decipher the functions of genes being only partially sequenced. One of the first commercial products of RNA silencing was tomato in which the target was to reduce the expression of these genes in the silenced plants meant that the tomatoes were firm after ripening and were not damaged by handling.
• RNAi is important for inhibition of gene expression at the post transcriptional level in eukaryotic cells.
• RNAi s is effective against parasites, so perhaps it can be used to silence parasitic genes.
Viruses have their say
An apparently peaceful meadow is actually the arena of fierce competition. The RNA Silencing is clearly a way of the plant to defend itself against pathogenic invaders, especially viral pathogens. As for viral pathogens, several of them have a way to fight back by suppressing the RNA silencing.
Plant viruses have elaborated a variety of counter defensive measures to overcome the host silencing response. One of these strategies is to produce proteins that target the cell autonomous or signalling steps of RNA silencing. It is not known whether a similar antiviral mechanism also operates in animal cells. Plants use RNA Silencing mechanism and produce short interfering RNA (SiRNA) molecules in a defense response against viral infection. To counter this defense response viruses produces suppressor proteins which can block the host silencing pathway or interfere with its function in plant cells. The target for many viral suppressors and the mechanism by which they function in plant cells are still largely unknown.
Early evidence that viruses encode RNA Silencing suppressor proteins came from experiments in which Silenced transgenes in plants were reactivated after virus infection or after the introduction of genes encoding candidate suppressor proteins using virus vectors or additional transgenes [2]. Silencing suppressors have been identified from positive strand RNA viruses and DNA containing viruses [16]. As a group, the plant viral silencing suppressors are diverse in sequence and evolutionary origin. They are also functionally diverse, with some targeting cell autonomous steps and others targeting systemic signalling steps. Before focussing on specific silencing suppressors, it is important to consider the biological consequences of viral silencing suppression. At first glance, there would appear to be a conflict. On the other hand, many viruses encode suppressor proteins that arrest cell autonomous or signalling steps, and many viruses encode functional suppressors that will trigger RNA silencing during infection. Since the discovery of the first viral suppressors of silencing in 1998, the literature has been satiated with reports of plant viral proteins that block silencing and with clues as to their mechanism of action [9,14].
Suppressor and small RNA function
The finding that primary siRNAs accumulate to high levels in the presence of certain suppressors, but that degradation of target RNAs is blocked, indicates that the primary siRNAs are not functional. In this regard, there are many reports of viral suppressors of silencing binding to small RNA duplexes, there by sequestering them and preventing their incorporation into the RISC effector complex (Figure 1) [8,12]. Indeed, sequestration of siRNA s has been proposed as a general mode of action for viral suppression of silencing. Viral suppressors can also alter the biochemical structure of siRNAs and this might well have a part in blocking their function. Previous studies have shown that plant endogenous small RNA, s and transgene siRNAs are methylated at their 3’ termini, an HUA ENHANCER 1 (HEN1) - dependent step in their biogenesis [10,18]. Methylation of viral siRNAs has also been demonstrated in plants infected with either DNA or RNA viruses, and several viruses and viral suppressors have been shown to interfere with both siRNA and miRNA methylation [4,1,18]. Furthermore, the virus, alteration of host miRNA accumulation and function is thought to underlie at least some symptoms of plant virus infection [7]. Although most such studies have focused on the role of viral suppressors, a recent study has shown that expression of other viral proteins can also affect miRNA accumulation and function [3]. Much of the early work on plant viral suppressors examined their role in transgeneinduced silencing. Those studies did not discriminate between primary and secondary siRNAs, and this led to confusion in the literature about whether a given suppressor did or did not block siRNA production. This seemingly contradictory results have now been resolved, with the findings that some viral suppressors (i,e. P1/Hc-Pro, P39, P19 siRNAs and leave primary siRNA accumulation unimpaired, where as other viral suppressors (i.e. P15 and P25) block accumulation of primary siRNAs [4,12,13]. The specific blockage in secondary siRNA accumulation might be produced simply by inhibiting primary siRNA function.
RNA silencing suppressors
Cucumoviral 2b
The 2b protein of cucumoviruses was recognized as a silencing suppressor at about the same time as P1/HC-Pro of potyviruses. The CMV 2b protein, a nuclear protein that is required for long distance movement of the virus, functions as the silencing suppressor (Hanako et al., 2013). Viral-suppressor protein 2b interact directly with components of the RISC machine, 2b interact with AGO1, by inhibiting its RNA-cleavage activity leading to the degradation of AGO1 (Figure 1). 2b specifically inhibits AGO1 cleavage activity in RISC reconstitution assays. In addition, AGO1 recruits virus-derived small interfering RNAs (siRNAs) in vivo, suggesting that AGO1 is a major factor in defense against CMV infection.
Potexviral P [25]
The P25 of the potexvirus, PVX is one of three cell-to-cell movement proteins (MPs) required for transport of virus from one cell to the next, the effects of P25 on cell autonomous and systemic silencing have been tested. Systemic silencing signal is a P25 – sensitive step and that the signal requires the transgene inducer pathway regardless of whether the inducer is a transgene or a replicating virus (Xi Q et al., 2006, Chiu et al., 2010) However the fact that a viral protein inhibits the pathway leading to systemic signalling strongly implies that the systemic arm of the silencing response is part of the antiviral defense mechanism.
HC-Pro
Helper component-Proteinase (a pathogenicity regulator of potyviruses) initially identified as a mediator of synergistic viral disease, acts to suppress the establishment of both transgene-induced and virus-induced gene silencing, and the Hc-Pro protein product is required for suppression. Hc-Pro binds to ds siRNA intermediates and have been suggested to function by sequestering ds siRNAs or by inhibiting their unwinding to ss siRNAs [22,23] Figure 1 Courtesy Trends in Microbiology, Vol.16. No.5. This discovery points to the role of gene silencing as a natural antiviral defense system in plants and offer different approaches to elucidate the molecular basis of gene silencing.
Tombusvirus P19
Since its discovery in the late 1980s, the status of the Tombusvirusencoded P19 protein (P19) changed from being thought obsolete to its identification a decade later as an important viral pathogenicity factor. The recent finding that P19 suppresses RNA interference (RNAi) by appropriating short interfering RNAs led to its widespread use as an RNAi-probing tool in various plant and animal models. Tombusvirus P19 is a protein encoded by tomato bushy stunt virus and related tombusviruses. Studies have demonstrated that P19 is an RNA silencing suppressor (RSS) in plant cells (Xiang et al., 2012). P19 was reported to suppress PTGS mainly along the vein tissue and in newly emerging leaves, whereas HC-Pro reversed PTGS in a nontissue- specific manner [16]. A study confirmed17 (i) P19 is a strong suppressor of PTGS (ii) P19 is a moderate suppressor of VIGS, and (iii) P 19 can also bind to ds siRNA by inhibiting their unwinding to ss siRNAs.
In the light of Co-evolution
Blast from the past
There are evolutionary interactions between species such that an evolutionary change in one species can prompt an evolutionary change in a species with which it interacts ecologically. Paleontological evidence already points strongly to an evolutionary “arm race” between herbivorous and carnivorous mammals during which size, speed and intelligence seemed to increase sequentially in various members of both groups.
Flor found 27 genes in the flax plant, Linum usitatissimum, that confer resistance against fungal rust pathogens while the pathogen in turn, had a similar number of genes allowing it to overcome resistance conferred by the host genes. In such cases, one can reasonably claim that an increased frequency of resistant mutation in the host will be followed by the selection of an increased frequency of one or more mutant genes in the parasite or pathogen that overcomes resistance.
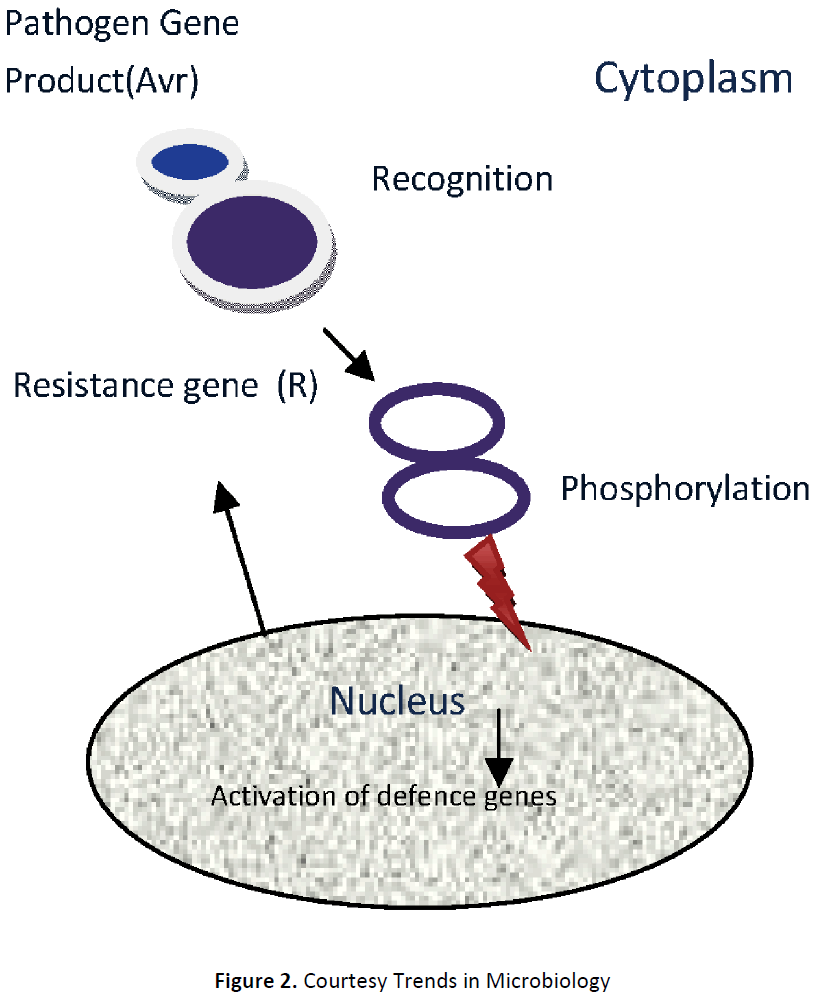
Gene-for-gene interactions in resistance (Figure 2. Courtesy Trends in Microbiology, Vol.16. No. 5).
Recognition of viral proteins by R proteins and induction of defense,
Plant Pathogen
R Gene Avr Gene
R Protein Avr Protein
CONCLUSION
Co-evolution must therefore be a common phenomenon since the intimate ecological relationships among the many species probably derive from co-evolutionary events in which adaptive changes in one species are followed by adaptive changes in others. There obviously has been and still is a great deal of interdependence among many life forms and theses cannot be fully understood except in an evolutionary frame work [11].
The plant has a resistance defensive mechanism as RNAi silencing, a mechanism that targets viral genomes and transcripts to degradation, several recent studies have revealed viral suppressors that target plant proteins and the possible actions that viruses take during their interference with the defense systems of the host: however many unanswered questions remain. For example, the type of proteolysis machinery used by PO to degrade its plant interactor AGO1 is a matter of debate and the mechanism by which V2 disrupts the RNAi- silencing system of the plant is unknown. The more we dig into the on-going battle between viruses and their hosts, the more we discover about the intriguing defense and counter defense that enable plants and viruses to coexist.
REFERENCES
- Akbergenov R, Ammour SA, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang, P, Gruissem W, Meins JrF, Hohn T, Pooggin M. Molecular characterization of geminivirus derived small RNAs in different plant species. Nucleic Acids Res. 2006; 34, 462–471.
- Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Smith, TH. & Vance, VB. A viral suppressor of gene silencing in plants. Proc Nat Acad Sci USA.1998; 95, 13079-13084.
- Bazzini AA, Hop HE, Beachy RN, Asurmendi S. Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Nat Acad Sci USA. 2007; 104, 12157– 12162.
- Csorba T, Aurelie B, Tamas D, Jozsef B. The p122 subunit of tobacco mosaic virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and microRNA-mediated pathways. J Virol. 2007; 81, 11768–11780.
- Ding SW, Vionnet O. Antiviral immunity directed by small RNA, s. Cell. 2007; 130, 413-426.
- Fagard M, Vaucheret H. Trans gene silencing in plants: how many mechanisms? Annual. Rev Plant Physiol and Plant Mol Biol. 2000; 51,167–194.
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Developmental Cell. 2003; 4, 205–217.
- Kurihara Y, Inaba N, Kutsuna N, Takeda A, Tagami Y, Watanabe Y. Binding of tobamovirus replication protein with small RNA duplexes. J Gen Virol. 2007; 88, 2347–2352.
- Li F, Ding SW. Virus counter defense: diverse strategies for evading the RNA- silencing immunity. Ann Rev Microbiol. 2006; 60, 503–531.
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 30-end uridylation activity in Arabidopsis. Current Biology. 2005; 15, 1501–1507.
- Loehle C, Pechmann JHK. Evolution: the missing in system ecology. American Naturalist.1988; 132, 884-899.
- Merai Z, Kerényi Z, Kertész S, Magna M, Lakatos L, Silhavy D. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J Virol. 2006; 80, 5747–5756.
- Mlotshwa S, Gail JP, Angela P, Endres MW, Junjie Li, Xumei Chen, Scot RP, Lewis HB, Vance V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. Arch PLoS ONE. 2008; 3, 1755.
- Silhavy D, Burgyan J. Effects and side- effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 2004; 9, 76–83.
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes and Development. 2006; 20, 759–771.
- Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc Nat Acad Sci USA. 1999; 96:14147-14152.
- Wenping Q, Jong-Won, Herman BS. Tombusvirus P19-Mediated Suppression of Virus-Induced Gene Silencing Is Controlled by Genetic and Dosage Features That Influence Pathogenicity. The Amer Phytopathol Soc. 2011; 15(3): 269–280.
- Yu B, Chapman EJ, Yang Z, Carrington JC, Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Letters. 2006; 580, 3117–3120.
- Shimura H, Masuta C, Yoshida N, Sueda Kae, Suzuki M. The 2b protein of Asparagus virus 2 functions as an RNA silencing suppressor against systemic silencing to prove functional synteny with related cucumoviruses. Virology. 2013. 442. 180– 188.
- Chiu MH, Chen IH, Baulcombe DC, Tsai CH (2010) the silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol Plant Pathol 11: 641–649. doi: 10.1111/j.1364-3703.2010. 00634.x.
- Xie Q, Guo HS (2006) Systemic antiviral silencing in plants. Virus Res 118: 1–6. doi: 10.1016/j. Virusres. 2005.11.012.
- Kasschau, K. D., Xie, Z., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A., & Carrington, J. C. (2003). P1/HC-Pro, a Viral Suppressor of RNA Silencing, Interferes with< i> Arabidopsis</i> Development and miRNA Function. Developmental cell, 4(2), 205-217.
- Arto J S, Balaji J, Kirsi L (2011). HC-Pro silencing suppressor significantly alters the gene expression profile in tobacco leaves and flowers. BMC Plant Biology. 11:68 doi:10.1186/1471-2229-11-68.
- Xiang Liu, Laurent H, Kuant T J (2012). Tombusvirus P19 RNA silencing suppressor (RSS) activity in mammalian cells correlates with charged amino acids that contribute to direct RNA-binding. Cell and Bioscience. 2. 41.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences