Phylogenetic Review of American Genus Macrocypraea(Mollusca, Caenogastropoda, Cypraeidae)BasedonPhenotypicCharacters
Luiz Ricardo L Simone*
1Department of Zoology Museum, Sao Paulo University, Brazil
- *Corresponding Author:
- Luiz Ricardo L Simone
Department of Zoology Museum, Sao Paulo University, Brazil,
Tel: +55 11 99676-2424
E-mail: lrsimone@usp.br
Received Date: April 05, 2021; Accepted Date: April19, 2021; Published Date: April 26, 2021
Citation: Simone LRL (2021) Phylogenetic Review of American Genus Macrocypraea (Mollusca, Caenogastropoda, Cypraeidae) Based on Phenotypic Characters. J Anat Sci Res Vol. 4 No.3:2.
Abstract
All Recent species of the American genus Macrocypraea are analyzed phylogenetically based on phenotypic features. The database was inserted in a wider phylogeny on Cypraeoidea previously published. The result is (((M. cervus (M. cervinetta (M. mammoth M. zebra))) Cypraea tigris) remaining Cypraeoidea). The monophyly of the genus us supported by four synapomorphies, and the proximity with C. tigris (among the anatomically known species) is demonstrated. The result is compatible with the subgeneric division Lorenzicypraea for M. cervus and Macrocypraea s.s. for remaining species.
Keywords
Anatomy; Morphology; Taxonomy; Systematics; Cypraeoidea
Introduction
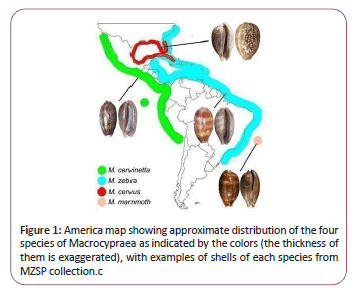
The genus Macrocypraea [1] (Caenogastropoda, Cypraeidae) only occurs in America (Figure 1). In Tropical and Subtropical Pacific coast occurs M. cervinetta [2]. Its counterpart in Atlantic coast is M. zebra [3], in such synonym, Cypraea exanthema [4], is the type species of the genus. Other two species have their distribution more restrict: M. cervus [5], occurring in Gulf of Mexico, Florida and Cuba; and M. mammoth [6], endemic from Trindade, a Brazilian oceanic island located approximately in middle of South Atlantic (Figure 1). This set of four species are all Recent species of the genus, that have other five fossil species from Eocene to Pleistocene of Florida and Central America [7,8]. The genus is usually divided into two subgenera, including the monotypic Lorenzicypraea [7] for M. cervus.
All Recent species of Macrocypraea have their anatomical details known, mainly by two recent papers [6,9], making a complementary phylogenetic study possible. A phylogenetic analysis based on phenotypic features is a good base for analyzing the taxonomy of its species, to determine which genera are closer related to it, and can be also used on a comparative basis with molecular analyses [10,11]. As a phylogeny of the Cypraeoidea based on phenotypy is available [9,12] the present paper has as objective to insert the surveyed data on four Macrocypraea species in that scenario [9] complementing that study, and saving long explanations on methodology in the present paper.
Materials and Methods
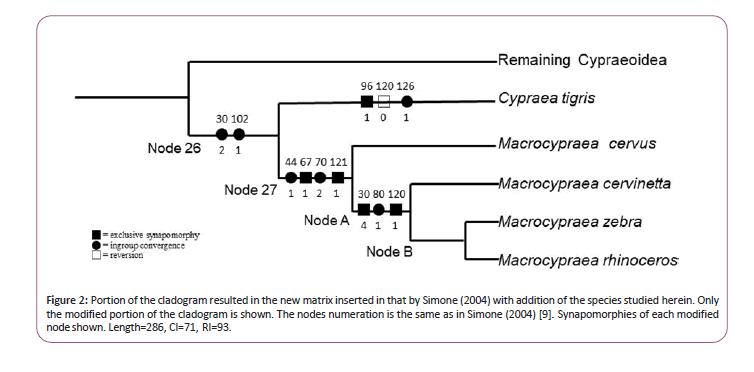
The list of material studied is already published and mostly consisted of fixed specimens deposited in museum’ collections [6, 9, 12]. The anatomical features of the species can be obtained as follows: for Macrocypraeazebra and M. cervinetta: [9]; for M. cervus and M. mammoth: [6]. Details of the phylogenetic analysis are found in [6,12,13], in such same methodologies and algorithms are applied herein. The set of characters in Table 1 are particularly based only on [9], which is more focused to cypraeids and already have two of the species (M. zebra and M. cervinetta) included. Data on M. rhinoceros and M. cervus [6] were analyzed and inserted in the data-matrix by Simone (2004: 173) [9]. Two lines were added to that matrix in Table 1 as well as two terminal taxa corresponding to both species. The new matrix was analyzed following the methods by [9]. The modified portion of that cladogram showing Macrocypraea and its sister-taxon, Cypraea tigris [3], is shown in Figure 2. The remaining cladogram is exactly the same as reported by [9] and is not illustrated.
Figure 2: Portion of the cladogram resulted in the new matrix inserted in that by Simone (2004) with addition of the species studied herein. Only the modified portion of the cladogram is shown. The nodes numeration is the same as in Simone (2004) [9]. Synapomorphies of each modified node shown. Length=286, CI=71, RI=93.
|
Taxon\character |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
12345 |
67890 |
12345 |
67890 |
12345 |
67890 |
12345 |
67890 |
12345 |
67890 |
12345 |
67890 |
12345 |
67890 |
12345 |
|
|
|
M. zebra |
11210 |
11010 |
00111 |
12100 |
01011 |
11014 |
00101 |
21211 |
00111 |
11121 |
11111 |
11012 |
11000 |
11112 |
11111 |
|
|
M. cervinetta |
11210 |
11010 |
00111 |
12100 |
01011 |
11014 |
00101 |
21211 |
00111 |
11121 |
11111 |
11012 |
11000 |
11112 |
11111 |
|
|
M. rhinoceros |
11210 |
11010 |
00111 |
12100 |
01011 |
11014 |
20101 |
21211 |
00111 |
11121 |
11111 |
11012 |
11000 |
11112 |
11111 |
|
|
M. cervus |
11210 |
11010 |
00111 |
12100 |
01011 |
11014 |
00101 |
21211 |
00111 |
11121 |
11111 |
11012 |
11000 |
11112 |
11111 |
|
Table 1: Matrix of characters added to the matrix by simone (2004) [9].
Results and Discussion
Using only the characters listed by [9], Macrocypraea cervus resulted as sister species to the species in the Macrocypraea branch (Figure 2) Node 27. The remaining three species resulted in a trichotomy (Node A). An additional analysis was performed with the addition of three characters, as follows: A) Anterior shell projection more elongated: exclusive to M. zebra and M. rhinoceros; B) Ocellated spots at lateral region of the shell: also exclusive to M. zebra and M. rhinoceros; C) Pallial papillae with base narrower than their middle region, with tip bearing 1-4 small projections: exclusive to M. zebra and M. rhinoceros. The addition of these 3 characters resulted in a different topology lacking polytomies: M. zebra and M. rhinoceros ended up in a same branch (Figure 2). Node B supported by the three above mentioned characters (A-C). In this case, the new cladogram in Figure 2 has length=86, CI=71, RI=93.
As reported above, the morphology-based phylogeny by [9] already contained M. zebra and M. cervinetta. The data on M. cervus and M. mammoth, joined to additional data on both anterior species, are in [6]. The presently obtained complementary cladogram (Figure 2). Reinforces the monophyly of Macrocypraea (Node 27). The node 27 is supported by four synapomorphies (character states as follows): 1) osphradium separated from posterior region of the gill (44/1); 2) pair of odontophore muscles mc comprising two bundles (67/1); 3) pair of m 11 with insertion surrounding ventral region of radular sac base (70/2); 4) bursa copulatrix located at middle level of pallial oviduct. In the topology presented herein, the first branch of Macrocypraea is M. cervus, separated from the other three species by three synapomorphies (Node A): 1) pallial papillae stubby, broad, with narrow base (30/4); 2) odontophore pair m7 originating at anterior border of ventral m4 branch (80/1); 3) bursa copulatrix U-shaped, bearing inner folds (120/1). The separation of M. cervus reinforces its separation in the subgenus Lorenzicypraea [7], in such it is type and single species. The remaining three species (Node A) represents the subgenus Macrocypraea s.s.
With the inclusion of the three extra characters discussed above, M. zebra and M. rhinoceros resulted in a proper node (Figure 2) Node B. Otherwise; both species resulted in a trichotomy with M. cervinetta. Moreover, Cypraea tigris, the type species of the genus (and, consequently, of the family and superfamily), resulted as the sister taxon to Macrocypraea s.s. node, a branch supported by two synapomorphies (Node 26):
1) Pallial papillae simple and long (30/2).
2) Presence of several transverse septa in the anterior duct of the digestive gland (102/1).
Conclusion
In some point of view, the subgenus Lorenzicypraea is valid and could be elevated to generic rank, a possibility that can be stressed in future wider revision. A different result, with M. zebra closer to M. cervus, rather than to M. cervinetta, was obtained in a molecular study. This competitor result must be analysed further deeper in future papers.
References
- Schilder FA (1930) Missbildungen an schalen der cypraeacea (Moll Gastr). Z Morph u Okol Tiere 19: 144-159.
- Kiener LC (1843) Genre Porcelaine. Spécies Général et Iconographie des Coquilles Vivante 1: 1-166, pls. 1-57.
- Carolus L (1758) By means of the three kingdoms of nature Created by Systema Naturae, secundum classes, ordines, genera, species, with the characters, the differences, synonyms, places. Lawrence Salviua: Stockholm 1: 1-824.
- Carolus L (1767) The three kingdoms of the nature, by a system of nature: Secundum classes, ordines, genera, species, with the characters, the differences, synonyms, places animal kingdom 1 and 2. Lawrence Salviua: Stockholm 1(2): 533-1327.
- Carolus L (1771). Mantissa plantarum altera generum editionis VI et specierum editionis II. Stockholm: Laurentius Salvius 1: 144-588.
- Simone LRL, Cavallari DC (2020) A new species of Macrocypraea (Gastropoda, Cypraeidae) from Trindade Island, Brazil, including phenotypic differentiation from remaining congeneric species. PLoS ONE 15(1): e0225963.
- Petuch EJ, Drolshagen M (2011) Compendium of Florida fossil shells. MdM Publisher 1: 1-412.
- Petuch EJ, Berschauer DP, Myers RF (2018) Jewels of the Everglades. The fossil cowries of southern California. San Diego Shell Club 1: 1-247.
- Simone LRL (2004) Morphology and phylogeny of the Cypraeoidea (Mollusca, Caenogastropoda). Papel Virtual 1: 1-185.
- Meyer CP (2003) Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biol. J. Linn. Soc 79: 401-459.
- Meyer CP (2004) Toward comprehensiveness: Increased molecular sampling within cypraeidae and its phylogenetic implications. Malacologia 46(1): 127-156.
- Simone LRL (2011) Phylogeny of the Caenogastropoda (Mollusca), based on comparative morphology. Arq. Zool. 42(4): 161-323.
- Simone LRL (2020). Bosquejos de Filogenia. Clube de Autores 1: 1-75.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences