ISSN : 0976-8505
Der Chemica Sinica
Nature and potential binding site prediction of covid-19 with Protease inhibitors using Pymol
Department of ITS College of Pharmacy, Muradnagar, Ghaziabad-201206, U.P. India
- *Corresponding Author:
- Shalini KS
Department of ITS College of Pharmacy
Muradnagar, Ghaziabad-201206, U.P. India
Tel: +919818830017
E-mail: shaliniksawhney@its.edu.in
Received Date: August 04, 2020; Accepted Date: August 12, 2021; Published Date: August 22, 2021
Citation: Shalini KS (2021) Nature and potential binding site prediction of covid-19 with Protease inhibitors using Pymol. Der Chem Sin Vol.12 No.9.
Abstract
Guillain Two recently reported crystal structures (PDB entries 6Y2E and 6LU7) of a protease from the SARS-CoV-2 virus, the infectious agent of the COVID-19 respiratory disease, has been investigated using pymol. Through this computational investigation the protease is predicted to display flexible motions in vivo which affect the geometry of a known inhibitor binding site. This opens new potential binding sites elsewhere in the structure. A database of the generated PDB files represents natural flexible variations on the crystal structures produced. This research article involves Docking and molecular interaction studies of COVID-19 with protease inhibitors using Pymol is aimed at identifying specific antiviral therapies for the treatment of COVID-19.
Keywords
SARS-CoV-2, COVID-19, Docking, protease inhibitors
Introduction
During end December 2019, it was observed that several people in Wuhan city of Hubei Province, China, suffered from SARS like pneumonia, which the World Health Organization (WHO) would later name COVID-19. The virus causing COVID-19 is a coronavirus with the taxonomic identifier SARS-CoV-2. [1] As per the WHO surveillance draft, in January 2020, any citizen in transit from Wuhan city 14 days before the onset of the symptoms is assumed to be infected by COVID-19. WHO also circulated interim guidance for laboratories which do the testing for this newly emerged infection with prevention and control guidance. The COVID-19 virus was assumed to emerged in a bat and got subsequently transmitted to individuals via wild animal market & sea food. Throughout the world, there are shadowing borders to stop the spread of the unknown virus, although some nations stopped flights to and from China. Through the first week of the year 2020, 41 confirmed cases were COVID-19 positive, leaving one person dead and seven in critical care. This figure is continuously increasing on a daily basis. There were confirmed more than 2200 deaths, mainly in mainland China. On January 20, 2020, the National Health Commission of China confirmed the human-to-human transmission of this new coronavirus outbreak. Ten days later, WHO declared COVID-19 as a Public Health Emergency of International Concern (PHEIC). COVID-19 symptoms include dry cough, fever, malaise, dry cough, respiratory distress and shortness of breath. COVID-19 is a member of Betacoronaviruses, like the earlier human coronaviruses SARS and MERS. There are now seven different strains of Human coronaviruses (HCoVs), namely, 229E and NL63 strains of HCoVs ( Alphacoronaviruses ), OC43, HKU1, SARS, MERS, and COVID-19 HCoVs ( Betacoronaviruses). The widely known strains of coronaviruses are SARS and MERS HCoV. Each has caused about 800 deaths. According to WHO, the mortality rates for SARS and MERS HCoV are 10% and 36%, respectively while COVID-19 has a 2% mortality rate [2].
Several crystal structures of proteins from SARS-CoV-2 have been found and made available through the Protein Data Bank (PDB). We can get a detailed knowledge about structure through the arrangement of atoms which makes up a protein complex from these crystal structures. This gives us all possible conformations of the protein/ protein complex. We also get natural dynamics about protein function.
Kern [3] describes natural dynamics as a property which directly affects the geometry, like, an enzyme active site. Methods like elastic network modelling, geometric simulation of flexible motion and rigidity analysis give the information about the low-frequency motions of a protein structure, largeamplitude, at the computational cost of molecular dynamics (MD) approaches. [4] These approaches are corresponding to computational MD studies and experimental biophysical/ biochemical studies of protein behaviour. Modern investigations has shown that the motions seen by geometric simulation bear a resemblance to the dynamics studied from MD trajectories[5]; the flexible motions can couple to enzyme active site geometry; and the structure produced from geometric simulation of a large amplitude motion is used as feedback for further MD studies, as the geometric simulation retains the local bonding geometry and constraint network of the input crystal structure and forbids major bonding distortions and steric clashes [6].
The simulations reported suggest that the protease structure has the capacity of significant flexible motions which alter domain orientations and affect the geometry of an inhibitorbinding site. The inputs and outputs of the simulations, principally consists of PDB files representing flexible variations on the protease crystal structures.
Material and Methods
Crystal structures of a viral protease were downloaded from PDB entries 6Y2E (free protease) and 6LU7 (inhibitor bound). Download structure in PDB or mmCIF format. The downloaded structure contains coordinates for a single protein chain, and symmetry operations to generate copies making up the crystal structure. The structure was visualised in PyMOL. Generate symmetry mates.
Appropriate symmetry mate was selected to make up the homodimer which is indicated in the PDB entry (under Global Stoichiometry) as the biological entity of interest. Other symmetry copies were deleted. Alter chain ID(s) in the symmetry copy to give each chain a unique chain ID. Hydrogens are added at electron-cloud positions, structure is processed through MolProbity website and all recommendations for side chain flips are accepted. Hydrogenated structure in which all hydrogens have a serial number of 0 are downloaded.
Structure is visualised in PyMOL for final cleaning and preparation. Alternate conformations are removed (those with an altloc entry which is not a blank or an A). Water molecules are removed and extraneous heterogroups like glycerol, acetyl etc. The cleaned structure is saved; here, PyMOL renumbers the atoms in a sequence, so that hydrogens now have suitable serial numbers.
Covalent and noncovalent interactions are identified from the atomic geometry of the protein structure using SBFIRST. This produces lists of covalent bonds, labelled as being either rotatable or non-rotatable (such as the C-N bond in the peptide main chain); hydrophobic tether interactions, wherever two nonpolar sidechains are closely adjacent; and polar interactions, including salt bridges and hydrogen bonds.
Polar interactions are assigned energies between 0 and -10 kcal/mol based on the donor-hydrogen-acceptor geometry.
Rigidity was analysed with FIRST as a function of hydrogenbond energy cutoff. This analysis groups atoms into rigid clusters by matching degrees of freedom against constraints. Review constraints based on rigidity information.
Based on the rigidity analysis, a cutoff of -3.0 kcal/mol appeared appropriate for the geometric simulations, in line with previous studies [7,8].
At this stage, a small number of noncovalent constraints (one in 6Y2E, three in 6LU7) was identified as items of crystal packing. Normal mode analysis extract only the alpha carbon positions and run Elnemo pdbmat (generation of matrix) and diagstd (diagonalise matrix) functions. When these eigenvector/eigenvalue pairs are sorted by eigenvalue from lowest to highest, this study examines modes 7 to 16.
Geometric simulation of flexible motion. For each normal mode of interest (7 to 16), FIRST was run with a chosen energy cutoff (-3.0 kcal/mol) and the earlier identified lists of covalent and noncovalent bonding constraints, provided with normal mode eigenvector and a direction of motion along with a directed step size 0.01Å, and appealing the FRODA engine implemented in FIRST.
FRODA carries out a series of steps. For every step, all positions of atoms are moved along the normal mode direction.
Then, the local atomic geometry defined by the bonding constraints and steric exclusion is restored by a few cycles of iterative relaxation. When the number of steps are defined (here 100), a numbered “frame” is written out as a PDB file. Such frames series describes the trajectory of flexible motion.
FRODA continues each run until a user-defined. maximum number of steps (in this case 1500) or until “jamming” occurs where the constraints can no longer be satisfied, typically when the motion has led to severe steric clashes through the collision of two domains.
Results and Discussion
In The 6Y2E and 6LU7 crystal structures contain open coordinates for one chain (A) of the protease, and in case of 6LU7, a chain (C) represents the bound inhibitor.
Biological homodimer is made from the input structures for analysis and simulation which consist of chain A and a symmetry copy (B). Chain C is exactly like chain D.
There are no unequivocal hydrogens in the X-ray structure, which are added using MolProbity [9]. When hydrogens get added, then it is possible to identify the covalent bonds, polar noncovalent interactions and hydrophobic-tether interactions using SBFIRST.
Based on the donor-hydrogen-acceptor geometry, the polar interactions are rated with an effective energy between 0 and -10 kcal/mol.
Pebble-game rigidity analysis [10,13] seperates the structure into flexible regions and rigid clusters depending on the distribution of degrees of freedom.
This depends on the set of polar interactions, which is based on energy cutoff that excludes. By involving weaker polar constraints, one large rigid cluster spreads across both chains of the protease dimer.
As weaker constrains are further removed, elimination of constraints, the beta-sheet regions become completely flexible.
The mobility studies are carried out with a cutoff of -3.0 kcal/ mol, like previous studies on enzymes [5,7].
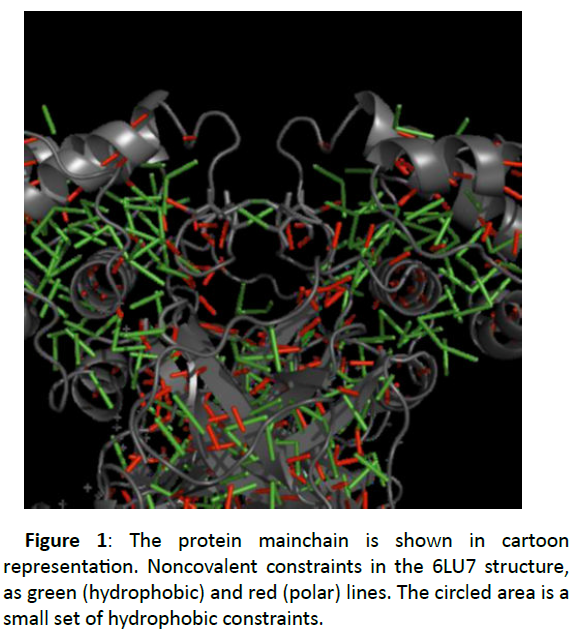
Even at this lower cutoff the structure is still constrained by noncovalent interactions (Figure 1).
Each folded domain is rich in hydrophobic interactions, at the interface region between two beta-sheet domains as the two chains make the homodimer, and collections of strong polar interactions constrain secondary structure (the backbone hydrogen bonds within alpha helices and beta sheets).
Figure 1 also shows a small set of hydrophobic constraints, one between the ALA 285 residues of the two chains, and one each between residue THR 280 of one chain and LEU 286 of the other. In the 6Y2E structure only the ALA 285 interaction is present. These are allocated between the N-terminal domains of chains A and B. In the 6Y2E structure, a single tether connects the side chains of the ALA 285 residues of both chains. In the 6LU7 structure, there are two extra tethers nearby, each connecting the side chains of the THR 280 residue of one chain and the LEU 286 of the other. If these tethers were retained, they would somewhat limit, the central cleft-opening motion.
Normal modes, which represent directions of motion, are taken from an elastic network model [11] representing protein as one site per residue and springs of uniform strength are placed between every pair of sites. In the 6LU7 structure, the amino acid residues of the bound inhibitor are comprised in the elastic network. Of the 3N normal modes of a structure with N residues, six modes of near-zero frequency represent the trivial rigid body motions of the structure, but the low-frequency modes from 7 upwards represent modes of flexible motion. The global geometry changes with minimum change in the local geometry, when the structure moves along a low-frequency mode direction making these directions of “easy” motion for the protein.
Lined projection of a protein structure lengthways a normal mode direction fastly introduce unphysical distortions into the structure. The geometric simulation approach implemented in FRODA [4,12], moves the structure along a normal mode direction, while maintaining steric exclusion and the bonding geometry and monovalent constraints of the input structure. These techniques give realistic flexible variations on the input structure and retain all-atom steric and bonding detail.
Protease structure is accomplished of substantial amplitudes of easy motion is shown by geometric simulations of the lowestfrequency nontrivial modes of motion (7 upwards).
The orientations of the alpha-helical domains relative to the beta-sheet domains can change through rotation and flexing about the interdomain “hinges”, and the beta-sheet domains are large enough to show bending and twisting motions among themselves.
Variations on a protein crystal structure are mostly applicable to structure-based drug design (SBDD) in two ways: a) flexible motion exposes clefts and possible binding sites not directly visible in the static crystal structure.
As the protein in vivo explores its flexible motion due to the Brownian-motion driving force of its solvent for e.g. cytosol, such latent sites establish valid target areas for inhibitors. b) global low-frequency motion couples to variations in the binding site/active site geometry [7,14].
Information about flexible deviation is very important for SBDD and/or fragment screening, as focussing on individual molecules that interact robustly with the binding site and can tolerate its flexibility is important in inclination to molecules that interact only with the crystal structure and not with its flexible variations.
A case of cleft opening in 6Y2E and 6LU7 structures shows some of the modes of motion (9- and 11+ in 6Y2E; 8+ and 11+ in 6LU7) have two alpha-helical domains moving separately from one another, exposing a cleft at the centre of the protein which is covered, narrow tunnel in the crystal structures.
The protein structures here are shown in a spacefilling (sphere) representation, as it is the exposed to new surface.
A more comprehensive sight of this opening cleft is shown in Figure 2. The residues are coloured to show the chemical character of the amino acid side chains.
Figure 2: Interdomain cleft opening (structure 6LU7, mode 11+, 1000th frame of geometric simulation). Protein structure is demonstrated as spheres and coloured by amino acid type. Green, aliphatic or aromatic residues; yellow, cysteine or methionine; red, hydroxyl (serine and threonine); magenta, acidic; purple, amidic; blue, basic. One end of the bound inhibitor is visible as grey spheres in the lower left. The exposed cleft is rich in basic, acidic and polar residues.
The portions of the alpha-helical domain surfaces that move to one side to open the cleft display hydrophobic residues, aliphatic or aromatic side chains.
The exposed surface area inside the cleft is lined with a series of basic residues, lysine and arginine, and is bordered by acidic residues aspartic and glutamic acids and polar residues threonine.
The basic and acidic residues show a network of inter- and intra-domain salt bridge interactions which stabilise the core of the homodimer in the crystal structures. An antagonist which can target this region (which has polar surface geometry with by flexible motion) could disturb the dynamics of the enzyme and interfere with its function.
The inhibitor is listed in the PDB as PRD_002214; it is a peptide-like inhibitor, with a central core of amino acids modified at either end with heterogroups, and was previously known to inhibit a protease of a feline coronavirus [15] In the 6LU7 structure, a binding site on the flank of each beta-sheet domain is occupied by the inhibitor N-((5-methylisoxazol-3- yl)carbonyl)alanyl-L-valyl-N~1~-((1R,2Z)-4-(benzyloxy)-4-oxo-1- (((3R)-2-oxopyrrolidin-3-yl)methyl)but-2-enyl)-L-leucinamide (chain C of the structure), hereinafter “N3”. In the 6Y2E structure the same binding cleft is empty, that is its geometry is likely to change in the course of flexible motion of the beta-sheet domain.
As the N3 inhibitor is deeply embedded in the binding site of the 6LU7 structure, opening and closing of cleft during protein’s natural motion is important for molecules to reach the cleft. By visual inspection, the region in the 6Y2E structure is considerably more closed as compared to 6LU7, with reliable intention that such closing andopening movements are fundamental to the protein.
Conclusion
Using pebble-game rigidity analysis, elastic network model normal mode analysis, and all-atom geometric simulations the flexibility and rigidity of two lately reported crystal structures (PDB entries 6Y2E and 6LU7) of a protease from the SARS-CoV-2 virus (the infectious agent of the COVID-19 respiratory disease) has been studied. This computer aided drug designing study of the viral protease tails protocols which is effective in investigating other homodimeric enzymes. The protease is foreseen to show flexible motions in vivo which affects the geometry of a known inhibitor binding site, e.g. through an opening/closing motion, and which open new possible binding sites somewhere else in the structure. A database of generated PDB files represens natural flexible variations on the crystal structures produced. This study may enlighten SBDD and fragment screening efforts to identify detailed antiviral investigations to treat COVID-19.
Acknowledgement
The author would like to thank the support given by the ITS College of Pharmacy, Muradnagar, Ghaziabad.
References
- WHO Naming the coronavirus disease (COVID-19) and the virus that causes it. (2020).
- Abdo A Elfiky, Ibrahim M. Ibrahim, Doaa H. Abdelmalek, Mohammed E. Elshahat (2020) COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect 80: 554-562.
- Henzler Wildman, K. and D. Kern (2007), Dynamic personalities of proteins. Nature, 450(7172): p. 964-72.
- Jimenez-Roldan, J.E., et al., Rapid simulation of protein motion: merging flexibility, rigidity and normal mode analyses. Physical Biology, 2012. 9(1)
- Wells, S.A, et al. ( 2015) Structure and Function in Homodimeric Enzymes: Simulations of Cooperative and Independent Functional Motions. Plos One, 10(8).
- Stephen A. W. Rigidity, normal modes and flexible motion of a SARS-CoV-2 (COVID-19) protease structure. Department of Chemical Engineering, University of Bath, BA2 7AY, UK. March 2020.
- Jones, H.B, et al. A complete thermodynamic analysis of enzyme turnover links the free energy landscape to enzyme catalysis. Febs Journal, 2017. 284(17): p. 2829-2842.
- Li, H.L., et al., Protein flexibility is key to cisplatin crosslinking in calmodulin. Protein Science, 2012. 21(9): p. 1269-1279.
- Williams, C.J., et al., MolProbity: More and better reference data for improved all-atom structure validation. Protein Science, 2018. 27(1): p. 293-315.
- Jacobs, D.J., et al., Protein flexibility predictions using graph theory. Proteins-Structure Function and Genetics, 2001. 44(2): p. 150-165.
- Suhre, K. and Y.H. Sanejouand, ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Research, 2004. 32: p. W610-W614.
- Wells, S.A., et al., Constrained geometric simulation of diffusive motion in proteins. Physical Biology, 2005. 2(4): p. S127-S136.
- Wells, S.A., J.E. Jimenez-Roldan, and R.A. Romer, Comparative analysis of rigidity across protein families. Physical Biology, 2009. 6(4).
- Jones, H.B.L., et al., Exposing the Interplay Between Enzyme Turnover, Protein Dynamics, and the Membrane Environment in Monoamine Oxidase B. Biochemistry, 2019. 58(18): p. 2362-2372.
- Wang, F., et al., Crystal Structure of Feline Infectious Peritonitis Virus Main Protease in Complex with Synergetic Dual Inhibitors. J Virol, 2016. 90(4): p. 1910-7.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences