Mutations of a Single Glycine in Vc-NhaP2, a Cation-Proton Antiporter in Vibrio cholerae, Confer the Ability to Exchange Li+ for H+
Carla B Schubiger1, Muntahi Mourin2, Craig T Resch2, Pavel Dibrov2* and Claudia C Häse1
1Department of Biomedical Sciences, Oregon State University, College of Veterinary Medicine, Corvallis, OR 97331, USA
2Department of Microbiology, University of Manitoba, Winnipeg, MB R3T 2N2, Canada
- *Corresponding Author:
- Pavel Dibrov
Department of Microbiology
University of Manitoba
Fort Garry Campus
Rm. 425 Buller Bldg
Winnipeg, Manitoba R3T 2N2
Canada
Tel: 1-204-474-8059
Fax: 1-204-474-7603
E-mail: Pavel.Dibrov@umanitoba.ca
Received date: May 30 , 2017; Accepted date: June 07, 2017; Published date: June 15, 2017
Citation: Schubiger CB, Mourin M, Resch CT, et al. Mutations of a Single Glycine in Vc-NhaP2, a Cation-Proton Antiporter in Vibrio cholerae, Confer the Ability to Exchange Li+ for H+. J Mol Biol Biotech. 2017, 2:2.
Abstract
The cation/proton antiporter Vc-NhaP2 from the important human pathogen Vibrio cholerae acts as a K+/H+ exchanger in vivo, contributing to the survival of V. cholerae at low pH and possibly enhancing the chances of the ingested V. cholerae cells to pass the acidic gastric barrier. Vc-NhaP2 is also able to exchange Na+ but not Li+ ions for H+. We attempted to identify the amino acids responsible for cation specificity of Vc-NhaP2 by limited alanine-scanning mutagenesis. Here we report a novel mutation, Gly159Ala, which enables Vc-NhaP2 to exchange Li+ for H+. Substitutions of Gly159 with Leu, Asp, or Lys, but not Ser had the same effect. Alanine substitution of the highly conserved Asn161 or Asp162 residues located nearby resulted in total inactivation of the antiporter. In the course of alanine-scanning mutagenesis, two distant residues, Asp273 and Leu289 that control the K+ /Na+ selectivity of Vc-NhaP2 were also identified. These findings support the idea of “ligand shading” in the active site of Vc- NhaP2, where different alkali cations are coordinated by overlapping but not identical sets of ligands, thus differently affecting the probability of protonation of the antiporter during the catalytic cycle. In a phylogenetic context, our results demonstrate one of the mechanisms underlying rapid divergent evolution of paralogous membrane transporters through the accumulation of seemingly insignificant single-point mutations (such as Gly-to-Ala) that might, nevertheless, have an immediate adaptive value.
Keywords
Mutagenesis; Novel function; NhaP type antiporter; Cation-proton antiport; Vibrio cholerae
Footnotes
Two first authors contributed equally to the study,
This research was supported by a grant from the National Institutes of Health 1 R21 AI109435- 01A1 (to CBS and CCH) and by grant # 227414-2012 from the Natural Sciences and Engineering Research Council of Canada (to CR, MM and PD). We would like to thank Dr. T. Nakamura (Niigata University of Pharmacy and Applied Life Sciences, Niigata, Japan) who kindly provided us with TO114 strain of E. coli. The abbreviations used are: PMF, the proton motive force; SMF, the sodium motive force; TMS, transmembrane segment; Ec-NhaA, Na+/H+ antiporter of NhaA type from Escherichia coli; Vc- NhaP, Na+/H+ antiporter of NhaP type from Vibrio cholerae; SDS PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; ΔpH, pH difference across the membrane; CPA, the extended monovalent cation-proton antiporter superfamily.
Introduction
Bacterial alkali cation/proton antiporters of the NhaP type form a surprisingly diverse group of membrane transporters [1] that contribute to the control of cytoplasmic pH and ion homeostasis by exchanging either monovalent cations such as sodium and/or potassium for protons [2-6] or K+ for Na+ through a heterological ion exchange [4]. The resulting transmembrane ion gradients can also serve as a direct energy source for a number of functions, such as the proton motive force buffering by ΔpK+ [7], Na+- substrate symports [8,9], or rotation of the Na+-dependent polar flagellar motors [10]. The first NhaP-type antiporter was identified in Pseudomonas aeruginosa in 1998 [2]. Presently, the NhaP family is considered as a part of the extended monovalent cation-proton antiporter 1 (CPA1) superfamily [1,11].
Of note, despite broad variations in Na+-K+ selectivity among the NhaP-type antiporters, Li+ is a poor substrate for most of them [1]. This seems paradoxical, as Na+ and Li+ ions have similarly small ionic radii (1.16 and 0.90 Å, respectively) while K+ is much bulkier (1.52 Å) and could be expected to require a larger coordination sphere [12]. Indeed, in the majority of Na+/H+ antiporters of different types studied up to date, Na+ and Li+ are equally good substrates while K+ is not transported at all [13]. Defying this general rule, Vc-NhaP2 from the human pathogen Vibrio cholerae acts as a K+(Na+)/H+ antiporter, with K+ as a preferred substrate both in vitro [4] and In vivo [6].
In vivo, Vc-NhaP2 acts as a K+/H+ exchanger, contributing to the survival of V. cholerae at low pH and possibly enhancing the chances of the ingested V. cholerae cells to pass the acidic gastric barrier [6]. In the absence of K+, Vc-NhaP2 was also able to exchange Na+ but not Li+ for H+ directly [4]. Ion competition assays in the inside-out membrane vesicles containing Vc-NhaP2, indicated that the antiporter can bind Li+ and exchange it for other alkali cations [4]. Nevertheless, direct Vc-NhaP2-mediated Li+/H+ exchange was undetectable [4]. Here we report an unusual mutation, Gly159Ala, which enables Vc-NhaP2 to exchange Li+ for H+ directly. Furthermore, substitutions of Gly159 with Leu, Asp, or Lys (but not Ser) also resulted in this new function, i.e., the direct Li+/H+ exchange. Alanine substitutions of the highly conserved Asn161 or Asp162 residues located nearby resulted in total inactivation of the antiporter. In addition, alanine substitution of two distant residues, Asp273 and Leu289, affected the K+/Na+ selectivity of Vc-NhaP2 without altering its Li+ discrimination.
Together, these findings support our previously proposed idea of “ligand shading” in the active site of Vc-NhaP2, where different alkali cations use overlapping but not identical sets of ligands, thus differently affecting the probability of protonation of the antiporter during the catalytic cycle [1]. Our findings also indicate that in the actual 3D-structure of Vc-NhaP2, these two distant groups of residues (in positions 159-162 and 273-289) might be located in close proximity, contributing to the coordination of translocated cations.
Experimental Procedures
Bacterial strains and culture conditions
The Na+/H+ antiporter-deficient strain of E. coli TO114 (nhaA::KmR, nhaB::EmR, chaA::CmR) was kindly provided by Dr. T. Nakamura (Niigata University of Pharmacy and Applied Life Sciences, Niigata, Japan) [3]. For cloning and plasmid construction, DH5α (supE44, hsdR17, recA1endA1, gyrA96, thi-1, relA1) (U.S. Biochemical Corp.) was used as the host. For the mutagenesis analysis the nhaP2 gene was amplified from the Vibrio cholerae strain O395-N1 [14]. For growth phenotype analysis a Na+/H+ antiporter-deficient (melBLid, ΔNhaB1, camR, ΔNhaA1, kanR, ΔlacZY, thr1) strain of E. coli, EP432 was used, kindly provided by Dr. E. Padan (Hebrew University of Jerusalem, Israel).
E. coli EP432 and TO114 cells were grown aerobically at 37◦C in LBK medium (modified Luria broth in which NaCl is replaced by KCl [15]) supplemented with 100 μg/ml ampicillin, 30 μg/ml kanamycin, 34 μg/ml chloramphenicol, 100 μg/ml erythromycin and 0.05% (w/v) arabinose.
Cloning and expression of Vc-NhaP2 mutant variants
The full-length Vc-nhaP2 gene was amplified from the genomic DNA of V. cholerae O395-N1 by high-fidelity PCR and cloned into the pBAD-TOPO vector (Invitrogen) under the arabinose-induced promotor (pBAD) as described in [4], but with a slight primer modification to eliminate the native stop-codon. The following primers were used for cloning: forward primer VcNhaP2expF 5 ’-GAGGAATAATAAGTGGACGCCGTTACGATTAAC-3’ and the new reverse primer VcNhaP2expR-STOPgone 5’-TTTCTCCGCGCCTTCTTGTAGCTC-3’. This modification extended the reading frame through the C-terminal tags (V5 and His) provided by the pBAD-TOPO vector. The resulting translation product, Vc-NhaP2, contained the entire ORF-encoded antiporter (including C-terminal V5 and 6×His tags) expressed from the arabinose-inducible pBAD promoter. Vc-NhaP2 mutant variants were constructed by site-directed mutagenesis using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). This method uses a high fidelity Pfu Fusion-based DNA polymerase, two synthetic oligonucleotide primers with the desired mutations and the supercoiled double-stranded DNA template. After amplification, nicks were sealed with the Pfu Fusion-based enzyme blend of the kit and the PCR product was digested with 2 μl of the restriction endonuclease DpnI at 37°C for 60 min to eliminate the methylated and hemimetylated template. Mutated ssDNA were either chemically transformed into XL10 Gold ultracompetent cells or electrocompetent E. coli KNabc as follows. 45 μl of XL10 Gold cells were thawed and gently mixed with 2 μl of β-ME and incubated on ice for 2 min. Then, 2 μl of DNA was added and again incubated on ice for 30 min, after which the cell mix was heat shocked for 30 sec at 42°C and then cooled for 2 min on ice before being mixed with 500 μl of SOC medium (Hanahan). Alternatively, the DNA was introduced into electrocompetent E.coli KNabc with a GenePulsar 1 (1,600 V, 25 mF and 400 Ohm; Bio-Rad), before being mixed with 1 ml of SOC. In both preparations, the cells were then incubated for 60 min at 37°C and shaking, before being selected on several pre-warmed LBK agar plates supplemented with 100 μg/ml ampicillin overnight at 37°C. Successful transformants were cultured, the plasmids isolated and submitted for sequencing at the Oregon State University Center for Genome Research and Biocomputing. For confirmation of the correct mutation and absence of amplification mistakes the entire gene and V5 tag were sequenced. Mutant variants were named for their targeted amino acid, followed by its location in the amino acid sequence and the amino acid it got mutated into, for example D162A means, that the aspartic acid located in position 162 of the amino acid structure was mutated into an alanine.
Growth phenotype assay
For growth analysis of mutant variants, LBB medium (non-cationic adjusted L broth) was supplemented with antibiotics, 0.0005% arabinose, and varying concentrations of KCl, LiCl or NaCl. The initial pH was adjusted to pH 6.0, pH 7.2, pH 8.0 and pH 8.5 by the addition of 60 mM Bis-Tris-Propane (BTP)-HCl. Cells were inoculated at a starting optical density at 600 nm (OD600) of 0.05 into 1.0 ml of liquid media placed in test tubes and grown at 37°C for 18 hours with vigorous aeration. Growth was then measured as OD600 by an Ultraspec 3000 spectrophotometer (Pharmicia Biotech). All experiments were repeated at least three times in triplicate.
Isolation of membrane vesicles and assays of antiporter activity
Inside-out membrane vesicles from TO114 cells transformed with the wild type (Vc-NhaP2) and Vc-NhaP2 mutant variants or pBAD24 (“empty” control) were isolated from cells grown in LBK medium at an OD600 of 1.0 to 1.2 and washed in buffer containing 140 mM choline-chloride, 10% (w/v) glycerol and 20 mM Tris-HCl, pH 7.5, and then resuspended in the same buffer containing 1 mM 1,4-dithiothreitol (DTT), 1 μg/ml pepstatin-A, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and approximately 5 mg/L DNase. The bacterial suspension was then passed through a French Press (Aminco), the unbroken cells were pelleted at 12,000×g for 10 min at 4°C, and the supernatant was ultracentrifuged at 184,000×g for 90 min at 4°C. The resulting membrane pellets were then resuspended and stored in the same buffer until assay for cation/proton antiport activity.
For measuring antiport activity, aliquots of vesicles (200 μg of protein) were added to 2 ml of buffer containing 140 mM choline chloride, 5 mM MgCl2, 10% (w/v) glycerol, 4 μM acridine orange and 50 mM BTP-HCl adjusted to the indicated pH. The cation/H+ antiport activity was measured using the acridine orange fluorescence quenching/dequenching assay. Respirationdependent generation of ΔpH was initiated by the addition of 20 mM Tris-D-lactate and the resulting quenching of acridine orange fluorescence was monitored in a Shimadzu RF-1501 spectrofluorophotometer (excitation at 492 nm and emission at 528 nm). Antiport activity was calculated based on its ability to dissipate the established ΔpH in response to the addition of 10 mM KCl. The antiport activities are expressed as percent restoration of lactate-induced fluorescence quenching. Each experiment was carried out in duplicate on at least six separate isolations of membrane vesicles.
Immunodetection of Vc-NhaP mutant variants in sub-bacterial vesicles
The TO114 cells were transformed with the expression plasmid pBAD-TOPO, containing wild type Vc-NhaP2 and Vc-NhaP2 mutant variants with the C-terminal V5 epitope for immunogenic detection. Membrane vesicles were isolated from each transformant as described above. For electrophoresis, 30 μl (~40 μg of total protein) of sample was loaded per lane. Proteins were resolved on a 10% polyacrylamide gel and blotted with monoclonal mouse-Anti-V5-HRP antibody (Novex) at a dilution of 1:3,000 in TBS containing 1% skimmed milk powder. For immune reactive signal detection, Pierce™ ECL Western Blotting Substrate was used.
Results
Selection of amino acids targeted for sitedirected mutagenesis
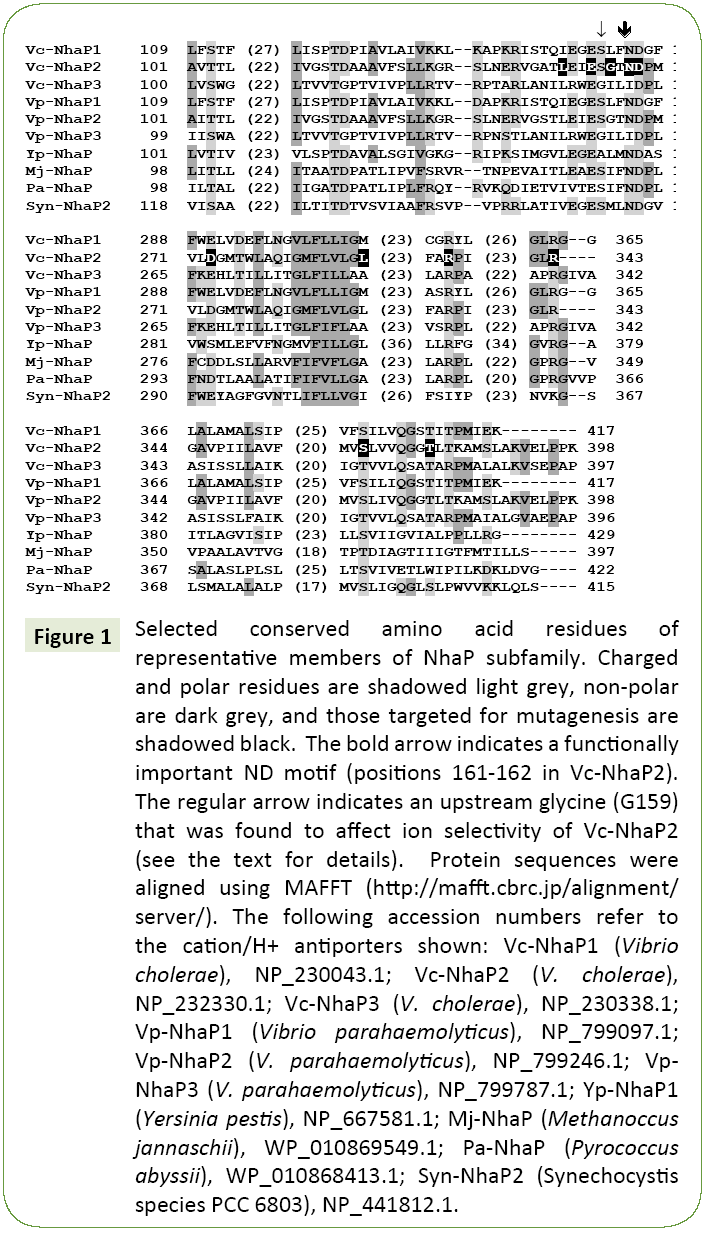
When crystallographic data are not available, mutational analysis remains a powerful tool of structural examination of membrane transporters [6,13-16]. Here we applied this approach to probe amino acid residues that might determine the ion selectivity of Vc-NhaP2. Sequence alignments of multiple NhaP-type antiporters revealed a significant number of highly conserved amino acid residues (Figure 1). Charged and polar amino acid residues associated with putative transmembrane segments are of special interest, as these residues typically form cationbinding pockets in Na+/H+ antiporters [16]. In particular, a ND motif (Asn158/Asp159) is present in NhaP1 from Methanococcus jannaschii [17] and in NhaP from Pyrococcus abyssi, where it was shown to be essential for antiport activity [18]. This ND motif is highly conserved in the aligned NhaP-type antiporters (Figure 1) as well as throughout the entire CPA1 family. It is also present in Vc-NhaP2, corresponding to Asn161 and Asp162 there, indicating the possibility of their involvement in cation binding. In this study, these two residues together with several others conserved amino acids were chosen for limited Ala-scanning mutagenesis. Overall, several conserved polar/charged as well as some nonpolar residues were selected. The various phenotypes resulting from alanine substitution were documented for each targeted residue (Table 1).
Figure 1: Selected conserved amino acid residues of representative members of NhaP subfamily. Charged and polar residues are shadowed light grey, non-polar are dark grey, and those targeted for mutagenesis are shadowed black. The bold arrow indicates a functionally important ND motif (positions 161-162 in Vc-NhaP2). The regular arrow indicates an upstream glycine (G159) that was found to affect ion selectivity of Vc-NhaP2 (see the text for details). Protein sequences were aligned using MAFFT (https://mafft.cbrc.jp/alignment/server/). The following accession numbers refer to the cation/H+ antiporters shown: Vc-NhaP1 (Vibrio cholerae), NP_230043.1; Vc-NhaP2 (V. cholerae), NP_232330.1; Vc-NhaP3 (V. cholerae), NP_230338.1; Vp-NhaP1 (Vibrio parahaemolyticus), NP_799097.1; Vp-NhaP2 (V. parahaemolyticus), NP_799246.1; Vp- NhaP3 (V. parahaemolyticus), NP_799787.1; Yp-NhaP1 (Yersinia pestis), NP_667581.1; Mj-NhaP (Methanoccus jannaschii), WP_010869549.1; Pa-NhaP (Pyrococcus abyssii), WP_010868413.1; Syn-NhaP2 (Synechocystis species PCC 6803), NP_441812.1.
Table 1 Effects of mutagenesis on the activity of Vc-NhaP2.
| Targeted Residue | Wild Type | Mutant Variant |
Observed Phenotype |
|---|---|---|---|
| 154 | L | A | Abolished antiport activity |
| 157 | E | A | Abolished antiport activity |
| 159 | G | A | Ion selectivity change |
| 159 | G | D | Ion selectivity change |
| 159 | G | K | pH profile and selectivity change |
| 159 | G | L | Ion selectivity change |
| 159 | G | S | pH profile change |
| 161 | N | A | Abolished antiport activity |
| 162 | D | A | Abolished antiport activity |
| 273 | D | A | K+/Na+ selectivity change |
| 287 | L | A | Abolished antiport activity |
| 289 | L | A | K+/Na+ selectivity change |
| 315 | R | A | Abolished antiport activity |
| 341 | G | A | Abolished antiport activity |
| 343 | R | A | Abolished antiport activity |
| 376 | S | A | Abolished antiport activity |
| 383 | T | A | Abolished antiport activity |
Mutations inactivating Vc-NhaP2 completely
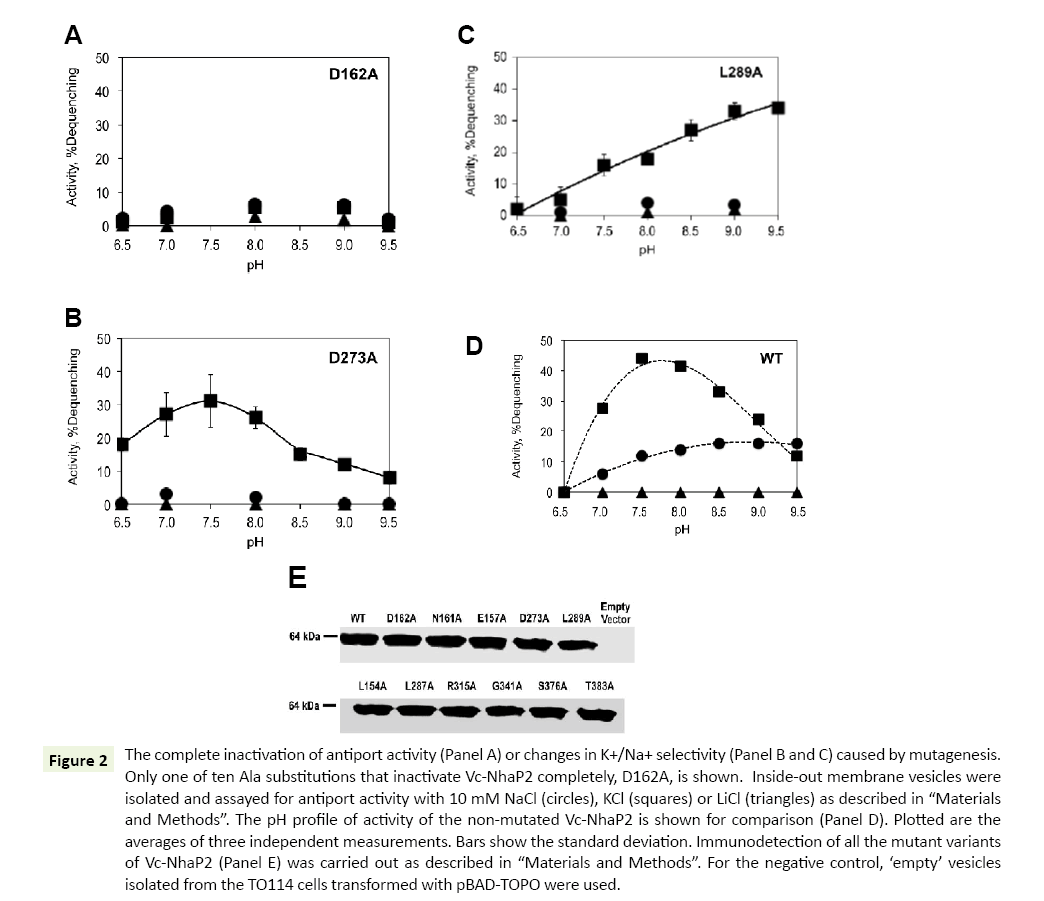
As shown in Table 1, ten of the analyzed alanine substitutions resulted in complete inactivation of Vc-NhaP2. This kind of phenotype is best exemplified by the Asp162Ala variant (Figure 2A). The other mutant variants with abolished antiport activity showed a similar pattern of activity with all alkali cations checked (data not shown). It has been reported that the mutation to alanine of Asp159 (an analog of Asp162 in Vc-NhaP2) and Asn158 (an analog of Asn161 in Vc-NhaP2), termed an ND motif, in Pa- NhaP resulted in complete loss of activity by the protein from P. abyssi [18]. In line with those findings, our results confirm that this ND motif, which is conserved in the CPA-1 family, is also critical for the antiport activity of Vc-NhaP2. Of note, residues with aliphatic, positively charged and polar side chains turned out to be critical for the activity of Vc-NhaP2, as well: Leu154 and Leu287, Arg315 and Arg343, Ser376 and Thr383 (Table 1). In all cases, the absence of activity cannot be attributed to affected expression levels and/or targeting of modified antiporter proteins, as the presence of the Vc-NhaP proteins in the membrane vesicles was confirmed by Western blotting (Figure 2E).
Figure 2: The complete inactivation of antiport activity (Panel A) or changes in K+/Na+ selectivity (Panel B and C) caused by mutagenesis. Only one of ten Ala substitutions that inactivate Vc-NhaP2 completely, D162A, is shown. Inside-out membrane vesicles were isolated and assayed for antiport activity with 10 mM NaCl (circles), KCl (squares) or LiCl (triangles) as described in “Materials and Methods”. The pH profile of activity of the non-mutated Vc-NhaP2 is shown for comparison (Panel D). Plotted are the averages of three independent measurements. Bars show the standard deviation. Immunodetection of all the mutant variants of Vc-NhaP2 (Panel E) was carried out as described in “Materials and Methods”. For the negative control, ‘empty’ vesicles isolated from the TO114 cells transformed with pBAD-TOPO were used.
Mutations suppressing the Vc-NhaP2-mediated Na+/H+ antiport
In NhaP-type antiporters, a negatively charged side chain is generally conserved in the position occupied by Asp273 in Vc- NhaP2, as well as a non-polar one at the position corresponding to Leu289, excluding two cases where polar methionine residues are present (Figure 1). Noticeably, in the V. cholerae paralogous variants, Vc-NhaP1 and Vc-NhaP3, there is either methionine or alanine in this position, which makes Leu289 an interesting residue to modify.
Mutations of Asp273 or Leu289 to alanine both resulted in a restrictive change in K+/Na+ specificity of Vc-NhaP2, converting it into an exclusive K+/H+ antiporter (Figure 2, Panels B and C, respectively). Whereas the Asp273Ala substitution changed the pH profile of remaining K+/H+ antiport only slightly, resulting in measurable activity at pH 6.5, where the wild type antiporter is inactive (compare Figure 2B to Figure 2D), the mutation of Leu289 to alanine caused a significant change in the pH profile (Figure 2C). Activity of the Leu289Ala mutant variant gradually rose as pH increased from 6.9 to 9.5, reaching the maximal levels ( approx 35% dequenching) at pHs above pH 8.5 (Figure 2C), while in case of the wild type NhaP2 it was below 20% at pH 9.5 (Figure 2D). Like the wild type Vc-NhaP2, both Asp273Ala and Leu289Ala variants showed absolutely no activity with Li+ at any pH examined (Figure 2, Panels B-D). Immunodetection again confirmed similar levels of these mutant variants in membranes of sub-bacterial vesicles (Figure 2E).
A new activity caused by Gly159Ala substitution: Direct Li+/H+ exchange
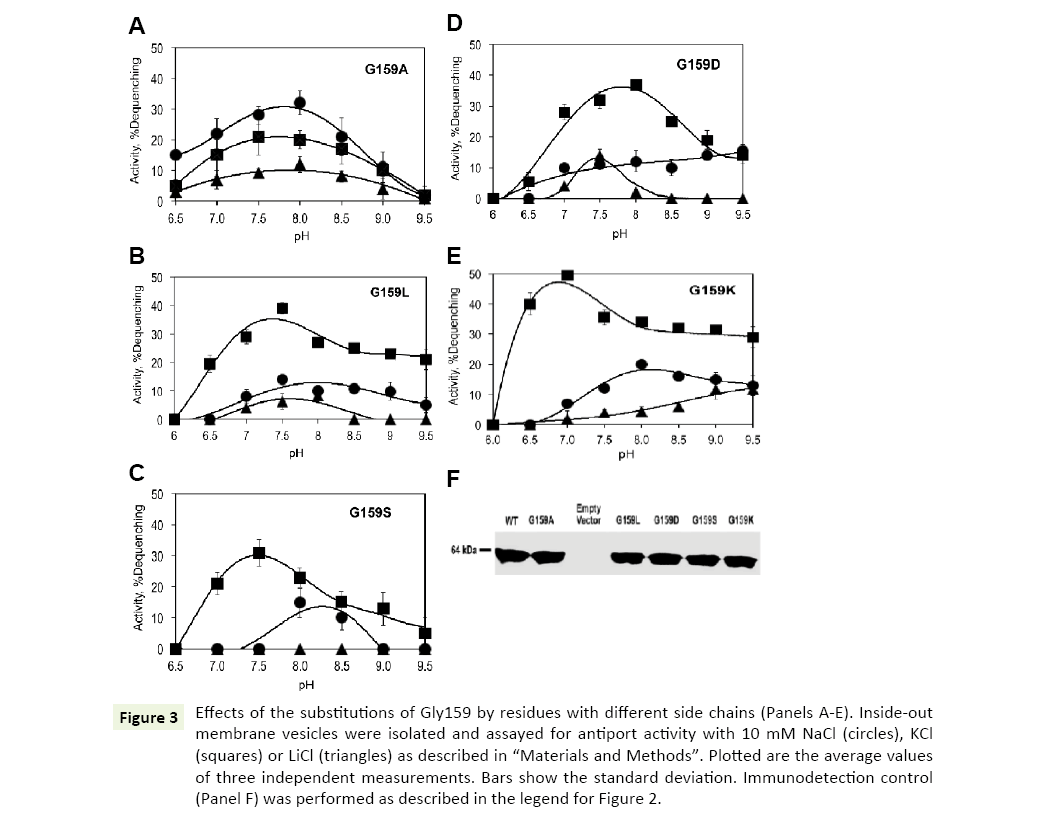
Gly159 is located in close vicinity of the highly conserved ND motif (Figure 1). Its substitution with alanine caused drastic changes in the ion selectivity of the antiporter. Interestly, introduction of a methyl group as the side chain at position 159 resulted in inverted ion preferences of Vc-NhaP2: now Na+/H+ antiport prevailed over K+/H+ antiport, reaching approx 30% dequenching at pH 8.0 (Figure 3A, circles). But, most importantly, the Gly159Ala variant also exhibited a new type of activity, which was absent in the wild type Vc-NhaP2, namely, direct Li+/H+ antiport. This modest but measurable Li+/H+ exchange peaked at approx 10% dequenching at pH 8.0 (Figure 3, triangles). Direct Li+/H+ exchange has never been observed in any of NhaP paralogues in V. cholerae [6].
Figure 3: Effects of the substitutions of Gly159 by residues with different side chains (Panels A-E). Inside-out membrane vesicles were isolated and assayed for antiport activity with 10 mM NaCl (circles), KCl (squares) or LiCl (triangles) as described in “Materials and Methods”. Plotted are the average values of three independent measurements. Bars show the standard deviation. Immunodetection control (Panel F) was performed as described in the legend for Figure 2.
Substitution of Gly159 with Aspartate, Lysine, Leucine and Serine
The presence of the bulkier alanine residue in place of glycine in the vicinity of a putative cation-binding residue altered the substrate selectivity of Vc-NhaP2. This raised a new question, namely whether a charged or a polar residue at this position would modify it even further, for example, by providing an additional ligand for Li+ binding. To test such a possibility, Gly159 was mutated to the negatively charged aspartate, positively charged lysine, polar serine and much bulkier non-polar leucine.
However, none of these substitutions altered K+/Na+ selectivity of Vc-NhaP, leaving K+ as the preferable substrate (Figures 3B-E). The Asp159, Lys159 and Leu159 variants were all able to mediate direct Li+/H+ exchange (Figures 3B, D, and E). Li+/H+ exchange in the Gly159Asp mutant variant showed a sharp maximum (approx 12% dequenching) at pH 7.5, whereas the variant possessing the positively charged lysine at this position, exhibited a gradual rise of Li+/H+ exchange from pH 7.5 to the maximal activities (approx 10% dequenching) at alkaline pH 9.0 and higher. Of note, the pHcontrol of both K+/H+ and Na+/H+ antiport was somewhat altered in this variant (Figure 3E, squares and circles). The bulkier side chain of leucine at the position 159 led to slightly decreased Li+/ H+ exchange activity compared to the Gly159Ala variant, showing >10% dequenching (Figure 3B, triangles). The Ser159 variant showed no Li+/H+ exchange, behaving similarly to the wild type NhaP2, with minor alkaline shifts in pH profiles of K+/H+ and Na+/ H+ antiport (Figure 3C). Therefore, introduction of a charge or polar group at the position 159 had no potentiating effect on Li+/ H+ exchange.
Again, immunodetection assays of Gly159Ala, Gly159Asp, Gly159Leu, Gly159Ser and Gly159Lys mutant variants in subbacterial vesicles confirmed that all of them were expressed and targeted to the membrane properly (Figure 3F).
Effect of Gly159Ala mutation on bacterial growth
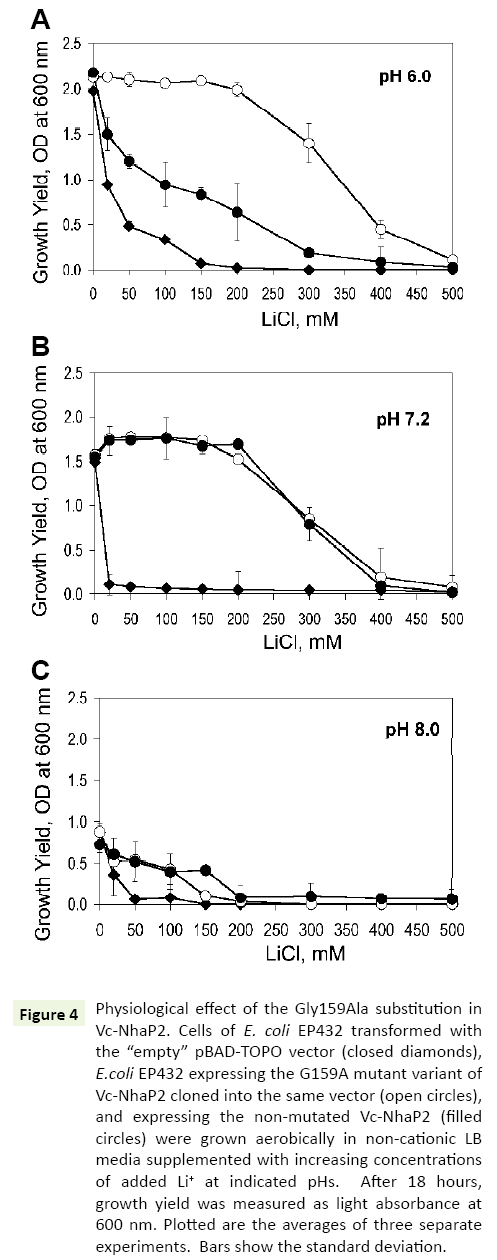
In the next series of experiments, a possible physiological effect of the G159A substitution was analyzed, by testing the possibility whether it enhances a protection of bacterial growth against toxic external Li+ In vivo. For these experiments, E. coli EP432, which is highly sensitive to environmental Li+ [13], was used. Similar to previous studies, at pH 6.0, its growth was markedly inhibited when 50 mM of LiCl was added to the medium and almost completely arrested at 150 mM (Figure 4A, filled diamonds). EP432 cells expressing wild type Vc-NhaP2 from a plasmid, exhibited a somewhat higher Li+ resistance (Figure 4A, filled circles). Such an effect was expected due to the ability of Vc-NhaP2 to exchange Li+ for Na+ (or K+) [4], which likely provided this protection against external Li+. As shown in Figure 4, transformation of E. coli EP432 with a plasmid containing the Gly159Ala variant resulted in markedly increased protection against high external LiCl concentrations. A normal growth yield was observed up to 200 mM of added LiCl, and a marginal growth ability was still registered in the presence of as much as 400 mM of LiCl (Figure 4A, empty circles). Remarkably, the Gly159 mutated variant of Vc-NhaP2 was able to exchange external protons for cytoplasmic Li+ ions, thus detoxifying the cells more efficiently.
Figure 4: Physiological effect of the Gly159Ala substitution in Vc-NhaP2. Cells of E. coli EP432 transformed with the “empty” pBAD-TOPO vector (closed diamonds), E.coli EP432 expressing the G159A mutant variant of Vc-NhaP2 cloned into the same vector (open circles), and expressing the non-mutated Vc-NhaP2 (filled circles) were grown aerobically in non-cationic LB media supplemented with increasing concentrations of added Li+ at indicated pHs. After 18 hours, growth yield was measured as light absorbance at 600 nm. Plotted are the averages of three separate experiments. Bars show the standard deviation.
However, at pH 7.0 and 8.0, wild type Vc-NhaP2 was as effective in conferring Li+ resistance as its Gly159Ala variant (Figures 4B and 4C, filled and empty circles). Such a phenotype was not surprising, because Vc-NhaP2 is an electroneutral cation-proton antiporter, and the transmembrane pH difference (ΔpH) is the sole driving force for the exchange of any substrate cation for proton [4]. Therefore, direct Li+/H+ exchange through the Gly159Ala variant, was expected to occur (and contribute to the overal Li+ resistance) only at sufficiently acidic external pH, where sizable ΔpH exists on the cell membrane [13].
Discussion
The data reported in this communication offer several new clues about the functional organization of the NhaP-type of antiporters, and in particluar the Vc-NhaP2 protein. First, while the Gly159, Asn161 and Asp162 residues are located in the putative transmembrane segment VI (TMS VI) of Vc-NhaP2, the Asp273 and Leu289 residues are both associated with the distant putative TMS X [6]. Nevertheless, mutation of either of them into alanine resulted in dramatic changes in the substrate specificity or abolished the antiport altogether (Asn161 and Asp162). Therefore, one may suggest that in an actual threedimensional structure, TMS VI and TMS X are close to one another, contributing to the formation of a cation-binding site of the antiporter. Such an arrangement indeed exists in the Na+/H+ antiporter from Escherichia coli, Ec-NhaA [15], belonging to the CPA-2 branch of the CPA family. In Ec-NhaA, unwound stretches of the discontinuous TMS IV and XI form a cation-binding cavity in the middle of the membrane [16]. Additional antiporters from CPA-2 also demonstrate a similar architectural theme. In Pa- NhaP, a Na+/H+ antiporter from a hyperthermophilic archaeon Pyrococcus abyssi, three negative/polar residues from three different TMSs (III, V and VI) participate in ion-binding [18,19]. Mj- NhaP from another thermophilic archaeon, Methanocaldococcus jannaschii, employs acidic residues from TMS V and VI to bind substrate ions [20]. The Asn161-Asp162 dyad, predicted to represent an ND motif that is conserved in all antiporters belonging to CPA-1 branch of the CPA family, predictably turned out essential for the activity of Vc-NhaP2, as well (Figure 2A). Of note, the electrogenic CPA-2 transporters have in the same place a conserved Asp-Asp motif with an additional negative charge [19,20]. Both residues were proposed to bind protons during the electrogenic antiport cycle [16], however N-to-D substitution in this motif did not affect Na+:H+ stoichiometry of Mj-NhaP [20]. Nevertheless, our alanine mutagenesis of Asn161 in Vc-NhaP2, together with the previously reported phenotypes for Mj-NhaP [20], indicates that the side chain of asparagine is still involved in ion translocation, albeit indirectly. In contrast, the negatively charged component of the motif (corresponds to Glu162 in Vc-NhaP2) seems to coordinate translocated cations directly in the archaeal NhaP-type antiporters mentioned above [18-20]. Therefore we suggest that Glu162 of the ND motif of Vc-NhaP2 is most probably directly involved in the coordination of translocated cations. However, further experiments will be needed to verify this prediction.
Another remarkable result was the acquisition of a new activity, namely direct Li+/H+ antiport, as a consequence of the substitution of Gly159 (located in the vicinity of the conserved Asn161-Asp162 motif) by another neutral, negatively or positively charged residue (Figure 3). To the best of our knowledge, mutations of aliphatic residues having such an effect have never been reported before. These findings are in accord with our previously suggested idea of “ligand shading” in the active site of Vc-NhaP2, where different alkali cations use overlapping but not identical sets of ligands, thus differently affecting the probability of protonation of the antiporter during the catalytic cycle [1]. As the electrical properties of the side chain were not important for this effect, one may conclude that the substitutions of Gly159 affected the delicately balanced overall conformation of the ion-coordinating pocket by insertion of a bulky radical in place of single –H rather than providing or eliminating ligands for coordination of substrate cations. As the result, binding of Li+ by the mutated Vc-NhaP2 did no longer prevent its protonation and, therefore, allowed for the direct Li+/H+ exchange.
In all tested substitutions of Gly159, the acquired Li+/H+ antiport remained minor compared to the K+/H+ and Na+/H+ antiport mediated by the mutated Vc-NhaP2 (Figure 3). Thus it was important to determine, whether such a modest additional activity brought into existence by a “neutral” mutation of a single glycine into alanine, could be of any physiological significance. It turned out that the ability of the mutated Vc-NhaP2 antiporter to exchange Li+ for H+ directly resulted in a considerably enhanced lithium resistance of the test strain, E. coli EP432, expressing the Gly159Ala variant of Vc-NhaP2 in acidic growth medium (Figure 4A). This result demonstrates how accumulation of seemingly insignificant mutations may drive a rapid divergent evolution of proteins encoded by paralogous genes, as exemplified by Vc-NhaP1, NhaP2 and NhaP3 co-existing in the membrane of V.cholerae [6].
Acknowledgments
This research was supported by a grant from the National Institutes of Health 1 R21 AI109435-01A1 (to CBS and CCH) and by grant # 227414-2012 from the Natural Sciences and Engineering Research Council of Canada (to CR, MM and PD). We would like to thank Dr. T. Nakamura (Niigata University of Pharmacy and Applied Life Sciences, Niigata, Japan) who kindly provided us with TO114 strain of E. coli.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Resch CT, Winogrodzki JL, Häse CC, Dibrov P (2011) Insights into the biochemistry of the ubiquitous NhaP family of cation/H+ antiporters. Biochem Cell Biol 89: 130-137.
- Utsugi J, Inaba K, Kuroda T, Tsuda M, Tsuchiya T (1998) Cloning and sequencing of a novel Na+/H+ antiporter gene from Pseudomonas aeruginosa. Biochim Biophys Acta 1398: 330-334.
- Radchenko MV, Waditee R, Oshimi S, Fukuhara M, Takabe T, et al. (2006) Cloning, functional expression and primary characterization of Vibrio parahaemolyticus K+/H+ antiporter genes in Escherichia coli. Mol Microbiol 59: 651-663.
- Resch CT, Patterson CT, Lind EJ, Quinn MJ, Dibrov P, et al. (2010) Putative Na+/H+ antiporter of Vibrio cholerae, Vc-NhaP2, mediates the specific K+/H+ exchange in vivo. Biochemistry 49: 2520-2528.
- Quinn MJ, Resch CT, Sun J, Lind EJ, Dibrov P, et al. (2012) NhaP1 is a K+(Na+)/H+ antiporter required for growth of Vibrio cholerae at low extracellular pH. Microbiol UK 158: 1094-1105.
- Mourin M, Schubiger CB, Resch CT, Häse CC, Dibrov P (2017) Physiology of the Vc-NhaP paralogous group of cation-proton antiporters in Vibrio cholerae. Mol Cell Biochem 428: 87-99.
- Brown II, Galperin MY, Glagolev AN, Skulachev VP (1983) Utilization of energy stored in the form of Na+ and K+ ion gradients by bacterial cells. Eur J Biochem 134: 345-349.
- Heyne RI, Vrij DW, Crielaard W, Konings WN (1991) Sodium ion-dependent amino acid transport in membrane vesicles of Bacillus stearothermophilus. J Bacteriol 173: 791-800.
- Häse CC, Barquera B (2001) Role of sodium bioenergetics in Vibrio cholerae. Biochim Biophys Acta 1505: 169-178.
- Atsumi T, McCarter L, Imae Y (1992) Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355: 182-184.
- Saier JRMH, Eng BH, Fard S, Garg J, Haggerty DA, et al. (1999) Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta 1422: 1-56.
- Gouaux E, MacKinnon K (2005) Principles of selective ion transport in channels and pumps. Science 310: 1461-5.
- Padan E, Venturi M, Gerchman Y, Dover N (2001) Na+/H+ antiporters. Biochim Biophys Acta 1505: 144-157.
- Mekalanos JJ, Swartz DJ, Pearson GD, Harford N, Groyne F, et al. (1983) Cholera toxin genes: Nucleotide sequence, deletion analysis and vaccine development. Nature 306: 551-557.
- Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S (1989) Deletion of antiporter in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ system(s). J Biol Chem 264: 20297-202302.
- Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, et al. (2005) Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435: 1197-1202.
- Goswami P, Paulino C, Hizlan D, Vonck J, Yildiz OZ, et al. (2011) Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1. EMBO J 30: 439-449.
- Wohlert D, Kuhlbrandt W, Yildiz O (2014) Structure and substrate ion binding in the sodium/proton antiporter PaNhaP. Elife 3: 03579.
- Calinescu O, Paulino C, Kuhlbrandt W, Fendler K (2014) Keeping it simple, transport mechanism and pH regulation in Na+/H+ exchangers. J Biol Chem 289: 13168-13176.
- Paulino C, Wohlert D, Kapotova E, Yildiz O, Kuhlbrandt W (2014) Structure and transport mechanism of the sodium/proton antiporter MjNhaP1. Elife 3: e03583.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences