ISSN : 2249 - 7412
Asian Journal of Plant Science & Research
Maize Germplasm Characterization Using Principal Component and Cluster Analysis
Solomon Mengistu*
Harar Biodiversity Center, Ethiopian Biodiversity Institute, Addis Ababa, Ethiopia
Abstract
In Ethiopian Biodiversity Institute Gene bank, huge collections of maize germplasm are not yet characterized for the magnitude of genetic variability from each other. Though knowing the contribution of individual characters is essential to focus on particular characters in cultivar development. Hence, this experiment was conducted on 92 maize accessions which were not yet characterized and 2 local checks to estimate the magnitude of genetic diversity among the genotypes and to identify the major agro-morphological characters contributing for the observed variations. The experiment was arranged in Augmented Design in seven blocks at Arsi-Negele in the 2016 main cropping season. The characters used for analysis were days to flowering, plant height, ear height, ear per plant, days to maturity, ear length, kernel rows per ear, thousand grain weight and yield per plot. The 94 genotypes were grouped into four clusters where cluster I, II, III and IV comprised 30, 21, 23 and 20 genotypes respectively. Early matured and short genotypes were grouped in cluster IV, late matured in cluster II and high yielding and tall genotypes in cluster I. The principal component analysis indicated that the first principal component (PC1) had an eigenvalue of 4.4 and reflects 48.85% of the total variation, this represents the equivalent of two individual variables and the two variables that weighted higher than the other variables are plant height and ear length. The second principal component (PC2) was recorded eigenvalue of 1.63 and maintained 18.11% of the total variation and related to diversity among genotypes due to ear per plant (EPP). Moreover, principal components 3 to 9 were shown to have more than one eigenvalue, thus they represent equivalent of one individual variable each accounted for 0.98%, 0.78%, 0.68%, 0.35%, 0.15%, 0.03% and 0% respectively toward the variation observed among genotypes. The result ensures the existence of high genetic divergence among the studied maize genotypes.
Keywords
Maize; Germplasm; Quantitative characters; Variability; Principal component analysis; Cluster analysis
Introduction
Maize (Zea mays L.) is one of the popular crops grown in the world, ranking second to wheat and followed by rice [1]. It occupies an important position in the world economy as food, feed, and industrial grain crop. It is a staple food for several million people in the developing world where they derive their protein and calorie requirements from it. Maize is among the leading cereal crops selected to achieve food self-sufficiency in Ethiopia [2]. Although, improved cultivars have been largely included in the national extension package, the national average yield of maize is only 3.45 tons/ha, which is far below the world average of 5.5 tons/ha [3]. In any crop germplasm resource not only serves as a valuable source of useful genes but also provides a wide genetic variability. Bringing improvement over existing crop varieties is a continuous process in plant breeding. To achieve this objective, the breeder has to identify diverse parents having superior genetic variability for combining desirable characters. Hence, knowledge of sound genetic diversity is crucial for undertaking any recombination breeding program. Multivariate statistical techniques used to analyze multiple measurements on each individual and used in the analysis of genetic diversity. Among the multivariate techniques, Principal Component Analysis (PCA) and cluster analysis had been shown to be very useful in selecting genotypes for breeding program that meet the objective of a plant breeder, PCA may be used to reveal patterns and eliminate redundancy in data sets, as morphological and physiological variations routinely occur in crop species [4,5]. Cluster analysis is commonly used to study genetic diversity and for forming core subset for grouping accessions with similar characteristics into homogenous categories. Cluster analysis is frequently used to classify maize accessions and can be used by breeders and geneticists to identify subsets of accessions which have potential utility for specific breeding or genetic purposes [6].Therefore, the objective of this study was aimed to estimate the magnitude of genetic diversity among the maize genotypes and to identify the major agro-morphological characters contributing for the observed variations.
Materials and Methods
The study was conducted during the year 2016 at the experimental field of Arsi-Negele, Oromia Regional State, Ethiopia. It is located at 7°21’N 38°42’E and at an elevation of 1940 m.a.s.l. It has a chromic and pellic vertisol with PH value of 5-7. The annual rainfall of the location is measured 915 mm with 27 ± 0.30°C mean daily temperature. 92 maize accessions obtained from Ethiopian Biodiversity Institute and two local checks named as check 1 and 2 were grown at farm site. The ninety two maize accessions without replication along with two replicated checks were arranged in augmented design. Individual plot size measured 9 m × 1.5 m with 4 rows planted at a spacing of 75*30 cm. Recommended doses of fertilizers were applied. The other management operations were done timely and properly to raise the crop uniformly. Twenty randomly selected plants were used for recording observations on days to flowering, plant height, ear height, ear per plant, days to maturity, ear length and kernel rows per ear, thousand grain weights, and yield per plot. The data collected for all quantitative characters were subjected to analysis of basic statistics, correlation, cluster and principal component analysis using the software statistical package for the social sciences (SPSS) 16.0 package [7].
Results
In the present study, genetic diversity was analyzed among 94 maize genotypes on the basis of 9 agro-morphological characters. The results of descriptive analysis, Ear Height (EH) showed the highest variation (35.52%) followed by number of ears per plant (30.39%) (Table 1). Conversely, the lowest variation was recorded from kernel rows per ear (6.01%) and days to maturity (9.61%).
| Characters | Mean | Minimum | Maximum | Range | SD | CV (%) |
|---|---|---|---|---|---|---|
| Days to flowering | 107.00 | 59.00 | 134.00 | 75.00 | 13.75 | 12.85 |
| Plant height (m) | 2.23 | 1.10 | 3.04 | 1.94 | 0.41 | 18.36 |
| Ear height(m) | 1.05 | 0.26 | 1.95 | 1.69 | 0.37 | 35.52 |
| Ear per plant | 1.95 | 0.00 | 3.10 | 3.10 | 0.59 | 30.39 |
| Days to maturity | 143.00 | 95.00 | 170.00 | 75.00 | 13.75 | 9.61 |
| Ear length(cm) | 15.13 | 10.70 | 19.00 | 8.30 | 1.80 | 11.90 |
| Kernel rows per ear | 12.28 | 9.80 | 14.40 | 4.60 | 0.74 | 6.01 |
| Thousand grain weight (g) | 338.37 | 196.00 | 504.00 | 308.00 | 54.47 | 16.10 |
| Yield (kg/plot) | 5.78 | 3.37 | 7.69 | 4.32 | 0.95 | 16.45 |
* SD = Standard deviation; CV = Coefficient of variation
Table 1: Basic statistics for various characters of maize genotypes.
Simple correlation coefficients confirmed that yield per plot recorded highly significant positive correlations among plant height, ear length, and thousand grain weight and maintained positive significant correlation among days to flower, ear height, and days to maturity and kernel rows per ear. Likewise, ear per plant was recorded insignificant negative correlations with days to flowering, plant height and ear height (Table 2). Similarly, found highly significant positive correlation of grain yield with cob diameter and thousand kernels weight and significant positive correlation with plant height [8].
| DF | PH | EH | EPP | DM | EL | KRPE | TGW | YPP | |
|---|---|---|---|---|---|---|---|---|---|
| DF | 1.00 | 0.59218** | 0.59323** | -0.092 | 1.00000** | 0.43338** | 0.18989 | 0.22982* | 0.25984* |
| PH | - | 1.00 | 0.96212** | -0.009 | 0.59218** | 0.68246** | 0.37618* | 0.47818** | 0.41423** |
| EH | - | - | 1.00 | -0.018 | 0.59323** | 0.62418** | 0.31869* | 0.41280** | 0.37949* |
| EPP | - | - | - | 1.00 | -0.09127 | 0.23183* | 0.13729 | 0.17099 | 0.0399 |
| DM | - | - | - | - | 1.00 | 0.43338** | 0.18989 | 0.22982* | 0.25984* |
| EL | - | - | - | - | - | 1.00 | 0.42315** | 0.57258** | 0.45637** |
| KRPE | - | - | - | - | - | 1.00 | 0.36272 | 0.31602* | |
| TGW | - | - | - | - | - | - | - | 1.00 | 0.83037** |
| YPP | - | - | - | - | - | - | - | - | 1.00 |
*Significant at P = 0.05, ** Significant at P = 0.01, DF= Days to flowering, PH= Plant height, EH= Ear height, EPP= Ear per plant, DM=Days to maturity, EL= Ear length, KRPE= Kernel rows per ear, TGW= Thousand grain weight, and YPP= Yield per plot
Table 2: Phenotypic correlation coefficients for different traits on maize genotypes.
Principal Component Analysis
The nine components which had eigen values equal to or greater than one were retained as meaningful interpretation (Table 3). The principal component analysis indicated that the first principal component (PC1) had an eigenvalue of 4.4and reflects 48.85% of the total variation, this represents the equivalent of three individual variables and the three variables that weighted higher than the other variables are Plant height (PH), days to maturity and Ear Length (EL) (Table 4). The second principal component (PC2) recorded eigen value of 1.63 and maintained 18.11% of the total variation and related to diversity among genotypes due to Ear Per Plant (EPP). Moreover, principal components 3 to 9were shown to have more than one eigenvalue, thus they represent equivalent of one individual variable each accounted for 0.98%, 0.78%, 0.68%, 0.35%, 0.15%, 0.03% and 0% respectively towards the total variation.
| Eigenvalue | 4.40 | 1.63 | 0.98 | 0.78 | 0.68 | 0.35 | 0.15 | 0.03 | 0.00 |
| % of total variance | 48.85 | 18.14 | 10.93 | 8.63 | 7.59 | 3.85 | 1.66 | 0.35 | 0.00 |
| Cumulative variance % | 48.85 | 66.99 | 77.92 | 86.55 | 94.14 | 97.99 | 99.65 | 100 | 100 |
Table 3: Principle component analysis of different characters on maize genotypes.
| Character | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 |
|---|---|---|---|---|---|---|---|---|---|
| DF | 0.36 | -0.43 | 0.07 | 0.29 | 0.30 | 0.00 | -0.04 | 0.00 | 0.71 |
| PH | 0.43 | -0.09 | 0.06 | -0.27 | -0.38 | 0.22 | -0.06 | -0.73 | 0.00 |
| EH | 0.41 | -0.13 | 0.07 | -0.25 | -0.43 | 0.33 | 0.07 | 0.67 | 0.00 |
| EPP | 0.03 | 0.40 | 0.76 | 0.42 | -0.03 | 0.28 | 0.06 | -0.02 | 0.00 |
| DM | 0.36 | -0.43 | 0.07 | 0.29 | 0.30 | 0.00 | -0.04 | 0.00 | -0.71 |
| EL | 0.38 | 0.18 | 0.21 | -0.05 | -0.17 | -0.85 | 0.17 | 0.05 | 0.00 |
| KRPE | 0.24 | 0.27 | 0.18 | -0.62 | 0.66 | 0.11 | 0.00 | 0.03 | 0.00 |
| TGW | 0.32 | 0.44 | -0.31 | 0.23 | 0.00 | 0.01 | -0.73 | 0.09 | 0.00 |
| YPP | 0.30 | 0.38 | -0.48 | 0.27 | 0.10 | 0.18 | 0.65 | -0.06 | 0.00 |
Table 4: Principle component analysis of different characters on maize genotypes.
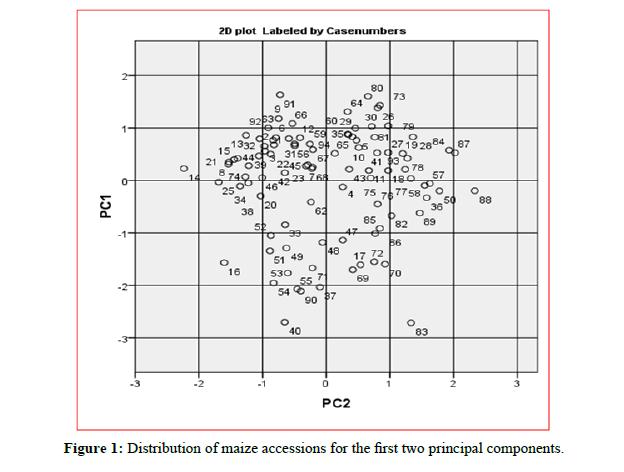
The PC3 showed high weights in ear per plant (EPP) and probably reflecting yield. The fifth principal component (PC5) kernel rows per ear (KRPE) had the largest weight, thus reflecting yield. The seventh principal component (PC7) showed high value on yield per plot (YPP). Eighth principal component (PC8) had a weighted high value of Ear Height (EH); this is probably reflecting the plant structure. Moreover, the ninth principal component (PC9) recorded the highest value on Days to Flowering (DF), thus reflecting flower development. In this study the principal component analysis categorized the total variance into nine (9) principal components and contributed maximum towards the total diversity (Figure 1). Similarly, reported the important contribution of the first PCs in the total variability while studying various traits. Principal component analysis (PCA) is usually used in plant sciences for the reduction of variables and grouping of genotypes [9,10]. Several authors suggested first principal component (PC) scores as input variables for the clustering process [11].
Figure 1: Distribution of maize accessions for the first two principal components.
Cluster Analysis
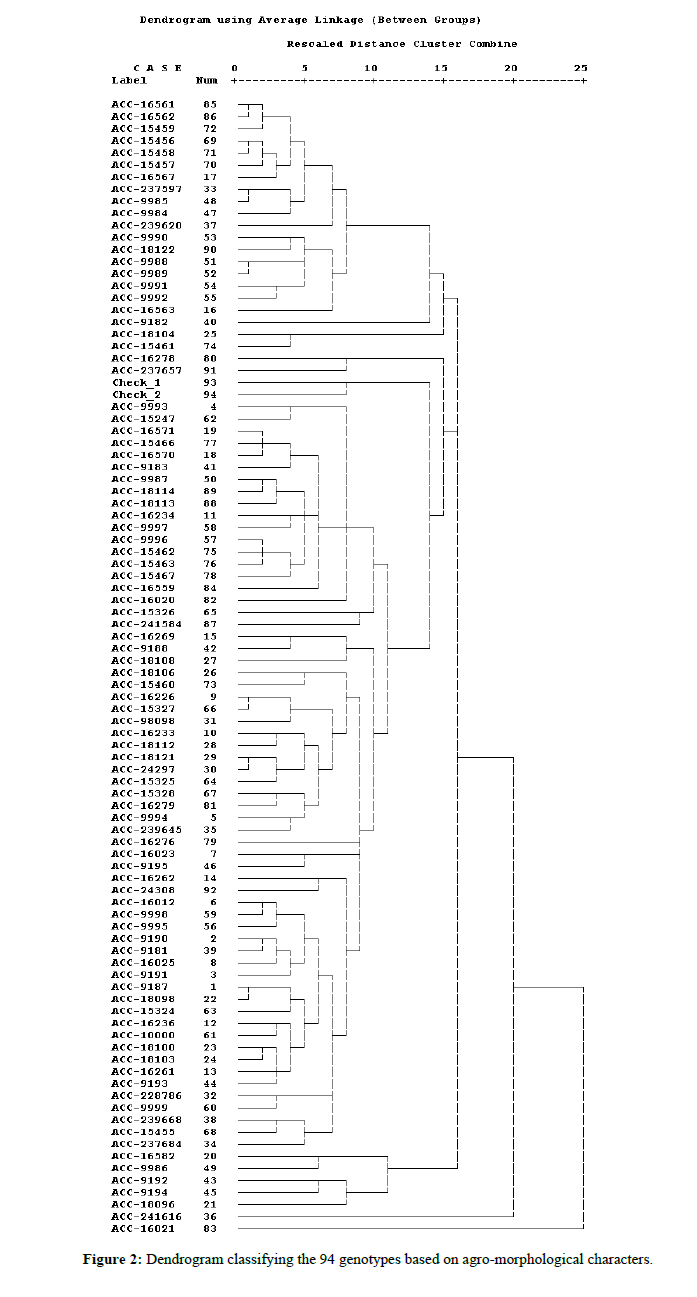
Clustering pattern of maize accessions under this experiment reveals that the maize germplasm showed considerable genetic diversity among them by occupying four different clusters (Table 5). These maize germplasm were grouped based mainly on day to flowering, plant height, ear height, ear per plant, days to maturity, ear length, kernel rows per ear, thousand grain weights and yield as variables. Ninety four maize genotypes were grouped into 4 clusters based on various agro-morphological characters. Cluster I to IV was comprised 30, 21, 23 and 20 maize genotypes respectively (Table 6). Thus, Cluster I was maintained maximum plant height(2.42 m), ear height (1.21 m), ear per plant (2.25), ear length (16.21 cm), kernel rows per ear (12.61) and yield (6.58 kg/plot). Cluster II showed late days to flowering (120.9 days) and maturing nature (156.9 days).Cluster III also maintained higher yield (5.88 kg/plot). Moreover cluster IV was showed relatively early maturing characters (133.5 days) but had minimum values of plant height (1.84 m), ear height (0.74 m), ear length (13.34 cm), kernel row per ear (11.82), and low yield (4.5 kg/plot). Similarly, hierarchical cluster analysis has been suggested for classifying entries of germplasm collections based on degree of similarity and dissimilarity [12]. A combination of cluster and principal component analysis has been used to classify maize (Zea mays L.) accessions [13].
| Cluster name | ||||
|---|---|---|---|---|
| Character | I | II | III | IV |
| DF | 109.2 | 120.9 | 99.7 | 97.5 |
| PH | 2.42 | 2.4 | 2.17 | 1.84 |
| EH | 1.21 | 1.2 | 0.97 | 0.74 |
| EPP | 2.25 | 1.75 | 1.71 | 2 |
| DM | 145.2 | 156.9 | 135.7 | 133.5 |
| EL | 16.21 | 15.56 | 14.9 | 13.34 |
| KRPE | 12.61 | 12.3 | 12.25 | 11.82 |
| TGW | 396.93 | 330.19 | 338.09 | 259.45 |
| YPP | 6.58 | 5.74 | 5.88 | 4.5 |
Table 5: Cluster means values for different agro-morphological characters of 94 genotypes.
| Cluster name | No. of genotypes | Name of accessions in each cluster | |||||
|---|---|---|---|---|---|---|---|
| I | 30 | ACC-9994 | ACC-16226 | ACC-16233 | ACC-16570 | ACC-16571 | ACC-18106 |
| ACC-18108 | ACC-18112 | ACC-18121 | ACC-24297 | ACC-9183 | ACC-9987 | ||
| ACC-9995 | ACC-9996 | ACC-9998 | ACC-9999 | ACC-15325 | ACC-15326 | ||
| ACC-15460 | ACC-15466 | ACC-15467 | ACC-16276 | ACC-16278 | ACC-16279 | ||
| ACC-16559 | ACC-241584 | ACC-18113 | ACC-237657 | Check_1 | Check_2 | ||
| II | 21 | ACC-9187 | ACC-9190 | ACC-9191 | ACC-16012 | ACC-16025 | ACC-16236 |
| ACC-16261 | ACC-16269 | ACC-18096 | ACC-18098 | ACC-18100 | ACC-18103 | ||
| ACC-228786 | ACC-237684 | ACC-239668 | ACC-9181 | ACC-10000 | ACC-15324 | ||
| ACC-15327 | ACC-15455 | ACC-24308 | |||||
| III | 23 | ACC-9993 | ACC-16023 | ACC-16234 | ACC-16582 | ACC-98098 | ACC-237597 |
| ACC-239645 | ACC-241616 | ACC-9188 | ACC-9192 | ACC-9194 | ACC-9984 | ||
| ACC-9985 | ACC-9986 | ACC-9997 | ACC-15247 | ACC-15328 | ACC-15462 | ||
| ACC-15463 | ACC-16020 | ACC-16561 | ACC-16562 | ACC-18114 | |||
| IV | 20 | ACC-16262 | ACC-16563 | ACC-16567 | ACC-18104 | ACC-239620 | ACC-9182 |
| ACC-9193 | ACC-9195 | ACC-9988 | ACC-9989 | ACC-9990 | ACC-9991 | ||
| ACC-9992 | ACC-15456 | ACC-15457 | ACC-15458 | ACC-15459 | ACC-15461 | ||
| ACC-16021 | ACC-18122 | - | - | - | - | ||
Table 6: Clustering pattern of the 94 accessions based on agro-morphological characters.
The tree diagram comprising 4 main cluster groups and each of them further subdivided into sub-clusters (Figure 2). The information regarding association among various traits is an important part for the initiation of any breeding programme and gives good chance for the selection of superior genotypes having desirable traits simultaneously [14].
Conclusion
Different methods are available to assess genetic variability among and within crop species. In the present study, principal component and cluster analysis techniques were employed to examine the amount of genetic variability present in a set of 94 maize accessions. Thus, it can be inferred from the present investigation that high level of genetic variability was present in agronomic and morphological traits like days to flowering, plant height, ear height, ear per plant, days to maturity, ear length, kernel rows per ear, thousand grain weight and yield per plot in the tested germplasm. Therefore, promising maize genotypes with more genetic divergences were identified. The identification of high levels of genetic variability during the current study could be indicated for germplasm characterization, conservation and further improvement in the maize breeding program.
References
- Vasal SK. Quality maize story. In: Improving Human Nutrition through Agriculture. The Roll of International Agricultural Research. A workshop hosted by IRRI, Philippines, Organized by International Food Policy Institute, Los Banos, USA. 2000.
- Bello OB, Malik SYA, Afolabi MS, Lge SA. Correlation and path coefficient hybrids analysis of yield and agronomic characters among open pollinated maize varieties and their F1 in a diallel cross. Afr J Biotech.2010, 9(18):2633-2639.
- Central Statistical Authority (CSA). Agriculture sample survey report on area and production for major crops for 2014/015.The FDRE Statistical Bulletin, CSA, Addis Ababa, Ethiopia. 2015.
- Mohammadi SA, Prasanna BM. Analysis of genetic diversity in crop plants-salient statistical tools and considerations. Crop Sci. 2003, 43:1235-1248.
- Adams MW. An estimate of homogeneity in crop plants with special reference to genetic vulnerability in dry bean Phaseolus vulgaris L. Euphytica. 1995, 26:665-679.
- Rincon F, Johnson B, Crossa J, Taba S. Cluster analysis. An approach to sampling variability in maize [Zea mays L.] accessions. Maydica. 1996, 4: 307-316.
- Levesque R. SPSS. Statistical package for the social sciences, version 16.0.SPSS Programming and data management. A guide for SPSS and SAS users, fourth edition, SPSS inc., Chicago III. 2007.
- Aliu S, Rusinvoci I, Fetahu S, Bislimi K. Morpho-physiological traits and mineral composition on local maize population growing in agro ecological conditions in Kosovo. Notulae Scientia Biol. 2013, 5(2):232-237.
- Mujaju C, Chakuya E. Morphological variation of sorghum landrace accessions on-farm in Semi-arid areas of Zimbabwe. Int J Bot. 2008, 4:376-382.
- Ali MA, Jabran K, Awan SI, Abbas A, Ehsanullah, et al. Morpho-physiological diversity and its implications for improving drought tolerance in grain sorghum at different growth stages. Australian J Crop Sci. 2011, 5(3):311-320.
- Mujaju C, Chakuya E. Morphological variation of sorghum landrace accessions on-farm in Semi-arid areas of Zimbabwe. Int J Bot. 2008, 4:376-382.
- Van Hintum TJL. Hierarchical approaches to the analysis of genetic diversity in crop plants. In: Hodgkin T, Brown AHD, Van Hintum TJL, Morales EAV (Eds), Core Collection of Plant Genetic Resources. John Wiley and Sons. 1995, 23-34.
- Crossa J, Delacy IH, Taba S. The use of multivariate methods in developing a core collection. In: Hodgkin T, Brown AHD, Van Hintum TJL, Morales EAV (Eds), Core collections of plant genetic resources. John Wiley & Sons, Chichester, UK. 1995, 77-92
- Ali MA, Nawab NN, Abbas A, Zulkiffal M, Sajjad M. Evaluation of selection criteria in Cicerarietinum L. using correlation coefficients and path analysis. Australian J Crop Sci. 2009, 3:65-70.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences