Intron-containing Housekeeping Genes as Useful Tools in Grapevine Virus Detection by PCR-based Protocols
Mihály Turcsán1,Tamás Deák2, Róbert Oláh1* and Ernő Szegedi1
1National Agricultural Research and Innovation Centre, Research Institute for Viticulture and Enology, Experimental Station of Kecskemét, 6000 Kecskemét, Katona Zsigmond út 5, Hungary
2Faculty of Horticultural Sciences, Department of Viticulture, Szent István University, 1118 Budapest, Villányi út 29-43, Hungary
- *Corresponding Author:
- Róbert Oláh
National Agricultural Research and Innovation Centre, Research Institute for Viticulture and Enology
Experimental Station of Kecskemét, 6000 Kecskemét
Katona Zsigmond út 5, Hungary
Tel: +36-76-506-430
E-mail: olah.robert@szbki.naik.hu
Received date: November 05, 2018; Accepted date: February 14, 2019; Published date: February 21, 2019
Citation: Turcsán M, Deák T, Oláh R, Szegedi E (2019) Intron-containing Housekeeping Genes as Useful Tools in Grapevine Virus Detection by PCRbased Protocols. Res J Plant Pathol Vol.2 No.1: 03.
Copyright: © 2019 Turcsán M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Mini-Review
Detection of viroids and RNA viruses infecting grapevines by PCR starts with RNA isolation, cDNA synthesis and validation of cDNA samples. Since the presence of residual plant genomic DNA may result in false positives during cDNA quality control, its complete removal that requires additional time and costs is basically important prior to cDNA synthesis. To overcome these difficulties we designed novel sets of primers specific for grapevine housekeeping genes that surround one or more introns. Therefore amplicons derived from remnant genomic DNA and cDNAs can be clearly distinguished by their sizes since cDNAs lack the intron sequences. Thus such primers used for internal controls make viroid and RNA virus detection by conventional PCR simpler since the time consuming DNA purification step can be avoided.
Grapevines like any other crop plants can be infected by several viroids and viruses which may cause serious economic losses. Until now approximately seven viroids and more than 70, mostly single-stranded RNA viruses have been identified in grapevines [1-5], but these numbers are continuously increasing since the introduction of novel deep sequencing technologies. At present no curative methods are available to control these diseases, thus their spreading can be prevented or at least delayed only by using pathogen-free propagating material. Healthy plants can be selected from existing plantations or produced by in vitro methods, like apical meristem and microshoot tip cultures alone or in combination with various physical or chemical treatments or through somatic embryogenesis [3,6]. The virus-free status of selected/produced plants should be confirmed by reliable diagnostic protocols, for which PCR-based methods are still most commonly used, although the recent development of nucleic acid sequencing methods provide several new alternatives to identify the complete virus populations (viromes) in plants [3,7].
The first steps of viroid and virus detection include isolation of high purity RNA followed by cDNA synthesis. For plant RNA isolation several excellent kits are available (SpectrumTM Plant Total RNA Kit by Sigma-Aldrich/Merck, RNeasy Plant Mini Kit by Qiagen and GeneJET Plant RNA Purification Mini Kit by Thermo Scientific), and conventional methods are published that usually use LiCl as selective precipitation agent for RNA [8,9]. To validate the efficiency of cDNA synthesis and to prevent false negative results various housekeeping genes are used which are stably expressed in the target plants/organs as internal controls. Since residual genomic DNA still present after LiCl precipitation [10] may cause false positive results during this step, the complete removal of genomic DNA is basically important. Thus the complete RNA purification is usually extended by several timeconsuming and costly steps, including enzymatic DNA digestion, additional organic extraction, repeated precipitation and washing which would be originally not necessary to detect viruses by RT-PCR or quantitative real-time-PCR. Furthermore, every additional steps of the isolation procedure diminish the theoretical sensitivity of the assay, which should be avoided especially when low virus titer has to be detected as it is frequently the case of propagation materials.
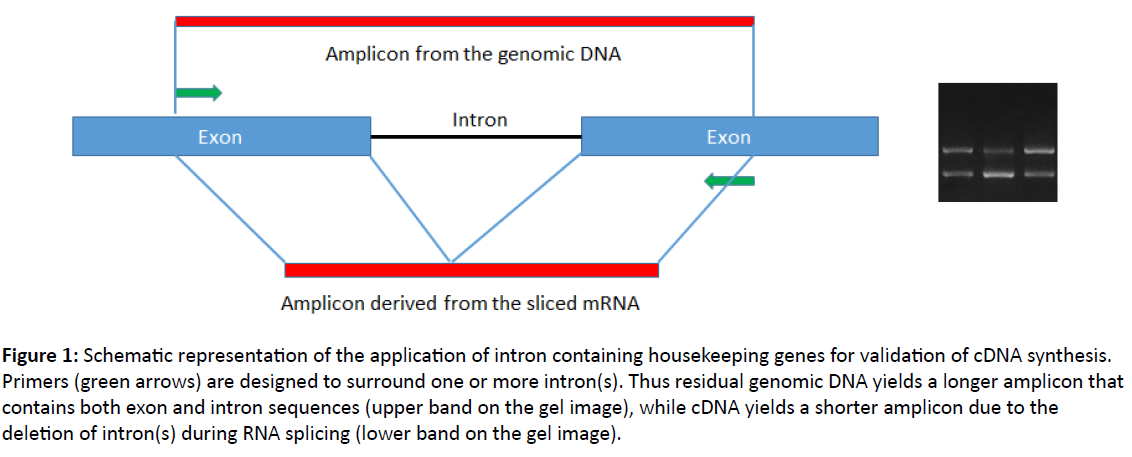
To eliminate these unnecessary steps of complete RNA purification we designed new sets of primers specific for four grapevine reference gene candidates phosphoenolpyruvate carboxylase, actin, tubulin and elongation factor 1-α (Table 1) [10,11]. In previous studies these genes showed stable expression under different conditions on the tested grapevine cultivars [12-16]. Primers were designed so that they surrounded one or more introns [10,11]. Thus amplicons derived from residual genomic DNA and those derived from cDNA can be clearly distinguished by their sizes on agarose gel, since intron sequences are deleted from cDNAs during RNA splicing (Figure 1). We have shown that the primers initiated the amplification of the expected regions from the genomic DNA samples of the 24 different grapevine cultivars tested, therefore they are suitable as reference gene specific primers for grapevines [10,11].
| Primer name | Target gene | Sequence (5’→3’) | No. of introns encompasssed | Annealing temperature (°C) | Length of the amplified fragment (cDNA/genomic DNA, bp) |
Reference |
|---|---|---|---|---|---|---|
| PepLfw PepLrev |
phosphoenolpyruvate carboxylase | CAAGGTGTCTGAGGATGACAAG GCTGTATCAACACGACGTAAGA |
3 | 54 | 717/2724 | [10] |
| PepSfw PepSrev |
phosphoenolpyruvate carboxylase | GTCCTTACAGCACATCCTACTC CCCACCCATCCAAGAAGAAA |
2 | 58 | 354/1597 | [10] |
| act-fw1 act-rev1 |
actin | GGCCGATACTGAAGATATCCAG ACCAGAATCCAGCACAATACC |
2 | 54 | 472/664 | [11] |
| tub-fw2 tub-rev2 |
tubulin | CACGATGCTTTCAACACCTTC CTTCATTGTCCAAGAGCACAG |
2 | 54 | 487/898 | [11] |
| elf-fw1 elf-rev1 |
elongation factor1 | GGGTAAGGAGAAGGTTCACATC TGCCTTGGAGTACTTTGGTG |
1 | 54 | 493/579 | [11] |
Table 1: Primers used in conventional PCR for internal controls to validate cDNA synthesis from grapevine RNA preparations.
Figure 1: Schematic representation of the application of intron containing housekeeping genes for validation of cDNA synthesis. Primers (green arrows) are designed to surround one or more intron(s). Thus residual genomic DNA yields a longer amplicon that contains both exon and intron sequences (upper band on the gel image), while cDNA yields a shorter amplicon due to the deletion of intron(s) during RNA splicing (lower band on the gel image).
Next we have tested RNA samples of grapevine leaves grown in vitro and in the field, petioles of field grown plants and cambial scrapings of wooden cuttings which are commonly used for virus detection assays. We have shown that each of the four tested genes uniformly expressed in the tested plant tissues/ organs of the 12 grapevine cultivars involved in this work. Our above studies show that the candidate gene sequences used for primer design are highly conserved among the different grapevine cultivars and these genes are stably expressed in different tissues/organs. Thus they can be reliably used as internal controls for the validation of cDNA synthesis in virus detection studies. These primers have been designed to aid quality control in cDNA synthesis for conventional PCR and not to be applied as an internal reference in quantitative real time PCR experiments.
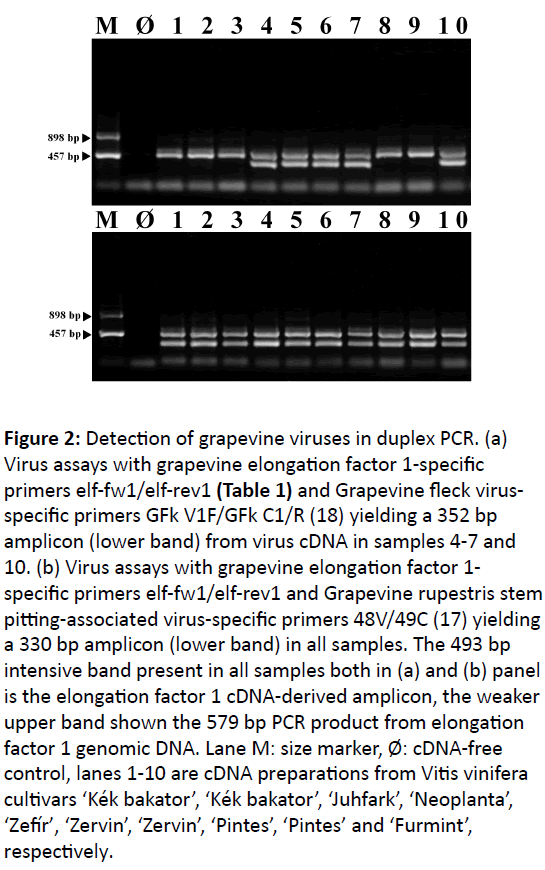
The suitability of phosphoenolpyruvate carboxylase genespecific primers PepSfw/PepSrev (Table 1) as cDNA quality controls in a Grapevine rupestris stem pitting-associated virus (GRSPaV) detection assay using the 48V/49C primers [17,18] was proven both in simplex and duplex PCR [10]. Grapevine cDNA samples validated with elongation factor 1 gene-specific primers elf-fw1/elf-rev1 (Table 1) were also appropriate to detect Grapevine fleck virus (GFkV) and GRSPaV (Figure 2) in duplex PCR. Further experiments should be carried out to test if the described reference gene-specific primers can be used in combination with pathogen specific primers in conventional duplex or multiplex PCR to detect the most important grapevine viruses [19]. To this end primers should not interact with each other, they should initiate DNA amplification under the same PCR conditions and amplicons derived from the host gene and pathogen-specific sequences should be clearly distinguished by their sizes on agarose gel. The principle described here surely can be applied for several other crops in viroid and RNA virus detection.
Figure 2: Detection of grapevine viruses in duplex PCR. (a) Virus assays with grapevine elongation factor 1-specific primers elf-fw1/elf-rev1 (Table 1) and Grapevine fleck virusspecific primers GFk V1F/GFk C1/R (18) yielding a 352 bp amplicon (lower band) from virus cDNA in samples 4-7 and 10. (b) Virus assays with grapevine elongation factor 1- specific primers elf-fw1/elf-rev1 and Grapevine rupestris stem pitting-associated virus-specific primers 48V/49C (17) yielding a 330 bp amplicon (lower band) in all samples. The 493 bp intensive band present in all samples both in (a) and (b) panel is the elongation factor 1 cDNA-derived amplicon, the weaker upper band shown the 579 bp PCR product from elongation factor 1 genomic DNA. Lane M: size marker, Ø: cDNA-free control, lanes 1-10 are cDNA preparations from Vitis vinifera cultivars ‘Kék bakator’, ‘Kék bakator’, ‘Juhfark’, ‘Neoplanta’, ‘Zefír’, ‘Zervin’, ‘Zervin’, ‘Pintes’, ‘Pintes’ and ‘Furmint’, respectively.
Acknowledgements
This work was supported by National Research, Development and Innovation Office (NKFIH) grants no. K-119783 and by GINOP grant no. 2.3.3-15-2016-00042.
References
- Maliogka VI, Martelli GP, Fuchs M, Katis NI (2015) Control of viruses infecting grapevine. Adv Virus Res 91: 175-227.
- Martelli GP (2014) Directory of virus and virus-like diseases of the grapevine and their agents. J Plant Pathol 96 (No. 1Suppl.): S1-S136.
- Meng B, Martellli GP, Golino DA, Fuchs M, eds. (2017) Grapevine Viruses: Molecular Biology, Diagnostics and Management. Springer International Publishing AG, Cham, Switzerland.
- Thompson JR, Fuchs M, McLane H, Celebi-Toprak F, Fischer KF, et al. (2014) Profiling viral infections in grapevine using a randomly primed reverse transcription-polymerase chain reaction/macroarray multiplex platform. Phytopathology 104: 211-219.
- Wilcox WF, Gubler WD, Uyemoto JK, eds. (2015) Compendium of Grape Diseases, Disorders, and Pests. 2nd edn, APS Press St. Paul MN USA.
- Wang MR, Cui ZH, Li JW, Hao XY, Zhao L, et al. (2018) In vitro thermotherapy based methods for plant virus eradication. Plant Methods 14:87.
- Czotter N, Molnar J, Szabó E, Demian E, Kontra L, et al. (2018) NGS of virus-derived small RNAs as a diagnostic method used to determine viromes of Hungarian vineyards. Front Microbiol 9:122.
- Gambino G, Perrone I, Gribaudo I (2008) A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem Anal 19: 520-525.
- White EJ, Venter M, Hiten NF, Burger JT (2008) Modified cetyltrimethylammonium bromide method improves robustness and versatility: the benchmark for plant RNA extraction. Biotechnol J 3: 1424-1428.
- Oláh R, Deák T, Turcsán M, Szénási M, Bordé Á, et al. (2017) Use of an intron containing grapevine gene as internal control for validation of cDNA synthesis in virus detection by RT-PCR. Eur J Plant Pathol 149:765-770.
- Szegedi E, Deák T, Turcsán M, Szénási M, Bordé Á, et al. (2018) Evaluation of intron-containing potential reference gene-specific primers to validate grapevine nucleic acid samples prepared for conventional PCR and RT-PCR. Vitis 57: 69-73.
- Borges AF, Fonseca C, Ferreira RB, Lourenco AM, Monteiro S (2014) Reference gene validation for quantitative RT-PCR during biotic and abiotic stresses in Vitisvinifera. PLoS ONE 9(10): e111399.
- Monteiro F, Sebastiana M, Pais MS, Figueiredo A (2013) Reference gene selection and validation for the early responses to downy mildew infection in susceptible and resistant Vitisvinifera cultivars. PLoS ONE 8(9): e72998.
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6: 27.
- Shinde MP, Upadhyay A, Sarika, Iquebal MA, Upadhyay AK (2016) Identification, characterization and expression analysis of ERF transcription factor VviERF073 and standardization of stable reference genes under salt stress in grape. Vitis 55: 165-171.
- Upadhyay A, Jogaiah S, Maske SR, Kadoo NY, Gupta VS (2015) Expression of stable reference genes and SPINDLY gene in response to gibberrelic acid application at different stages of grapevine development. Biol Plant 59: 436-444.
- Lima MF, Alkowni R, Uyemoto JK, Golino D, Osman F, et al. (2006) Molecular analysis of a California strain of Rupestris stem pitting-associated virus isolated from declining Syrah grapevines. Arch Virol 151: 1889-1894.
- Osman F, Leutenegger C, Golino D, Rowhani A (2008) Comparison of low-density arrays, RT-PCR and real-time TaqMan RT-PCR in detection of grapevine viruses. J Virol Meth 149: 292-299.
- Gambino G, Gribaudo I (2006) Simultaneous detection of nine grapevine viruses by multiplex reverse transcription-polymerase chain reaction with coamplification of a plant RNA as internal control. Phytopathology 96: 1223-1229.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences