Genetic Diversity and Evolutionary Analysis of Okra (Abelmoschus esculentus (L.) Moench) Germplasm Resources based on ISSR Markers
Ji Huang1#, Shucan Liu1#, Xinhong Guo1*, YanKe Zhao2, Meiling Li1, Cheng Zhang1, Jingjing Zhu1 and Jiazhuo Ye3*
1College of Biology, State Key Laboratory of Chemo/Biosensing and Chemometrics, Hunan University, Changsha, 410082, PR China
2Hunan University Library, Hunan University, 410082, PR China
3College of Electrlical and Information Engineering, Hunan University, 410082, PR China
#These authors contributed equally to this work
- *Corresponding Authors:
- Xinhong Guo
College of Biology
State Key Laboratory
of Chemo/Biosensing and Chemometrics
Hunan University, Changsha
410082, PR hina
Tel: 18711081973
E-mail: gxh@hnu.edu.cn - Jiazhuo Ye
College of Electrlical and Information Engineering
Hunan University, 410082
PR China
Tel: 18711081973
E-mail: yjz@hnu.edu.cn
Received date: September 14, 2017; Accepted date: September 14, 2017; Published date: October 03, 2017
Citation: Huang J, Liu S, Xinhong G, Zhao Y, Li M, et al. (2017) Genetic Diversity and Evolutionary Analysis of Okra (Abelmoschus esculentus (L.) Moench) Germplasm Resources based on ISSR Markers. Genet Mol Biol Res Vol. 1 No. 1:2
Abstract
Objective: Okra (Abelmoschus esculentus (L.) is a flowering plant in the mallow family, valued for its edible green fruits. The genetic diversity of Okra was analyzed to provide the theoretical basis for the collection, identification, protection and utilization of the germplasm resources of Okra plants.
Method: The genetic diversity among 34 accessions of okra from different regions of China was investigated using Inter-simple Sequence Repeat (ISSR).
Results: Out of the 128 repeatable bands detected, 90 bands were polymorphic. The percentage of polymorphic bands was 70.3%, with a mean of 7.6% of polymorphic bands per primer. With the 128 bands, the phylogenetic trees were constructed using the Unweighted Pair-Group Method with Arithmetic Average (UPGMA) cluster analysis. The 34 accessions were divided into two major groups in the evolutionary tree, and the two-dimensional matrix also confirmed this conclusion. The polymorphism information content (PIC) values varied from 0.4507 to 0.6801, indicating that the primers used were authentic.
Conclusion: The genetic distance among the sample accessions ranged from 0.6 to 0.94. The data showed that there was a high degree of genetic diversity among the okra accessions revealed by the ISSR markers. Our results confirm the potential value of genetic diversity preservation for future breeding of Okra.
Keywords
Okra; Genetic diversity; Intersimple sequence repeat (ISSR) markers
Introducción
Okra, a kind of annual herb, is a number of Malvaceae family, as well as is called as Abelmoschus esculentus, pepper and so on. It originated from East Asia and West Africa, spread later to tropical, subtropical, and warm temperate regions [1]. With the development of the technology, it was cultivated and distributed across the world. It is not considered to be a vital crop until more attention was paid to the improvement of genes, especially its rapid growth cycle, easy cultivation, protection against pests, high yield, and abundant nutritional materials [2]. As more studies on okra reported, it gets more attention owing to its features of high nutritional value, significant health and therapeutic effects function in the recent years [3]. The tender fruit pods of okra are rich in protein, unsaturated fatty acids, minerals, alkaloids, free amino acid, sticky slippery juice and other bioactive components [4], which can reduce blood sugar, protect the liver, cardiovascular and intestinal effects bringing excellent medicinal health effects in the condition of taking for a long periods of time. The leaf and seed of okra can be used as traditional medicines [5]. In addition, the beautiful flower of okra is also considered as ornamental plants [6]. Over the past decades, a series of achievements in cytology, introduction and cultivation techniques of okra is reported. However, there are few studies on the genetic diversity of germplasm resources, which is of significant importance for the identification and breeding [7]. Therefore, it is urgent to study the genetic diversity and genetic relationship of okra accessions in the identification, breeding and utilization of okra, which will provide references for the classification of okra [8].
The ISSR marker technique was proposed by Zietkiewicz [9]. This technique is widely used in studies on the genetic diversity of plants, for instance cotton [10], ryegrass [11], and ipomoea [12]. What’ more, it possesses advantages, such as good stability, high polymorphism, simple operation and time-consuming. The principle is to use Microsatellite DNA as a primer, and 2 to 4 random nucleotides were extended at the SSR sequence 3' or 5' termini. In the polymerase chain reaction (PCR), the primer caused specific sites annealing after amplified, which resulted in the repetitive sequence DNA fragments [13]. Multiple bands of the amplified ISSR region were resolved by agarose gel electrophoresis or PAGE electrophoresis. ISSR, a dominant marker, reveals some characteristics of the whole genome.
In this study, ISSR molecular marker technique was used to analyze the genetic diversity of 34 okra accessions and make some preliminary explorations for identification of okra germplasm.
Materials and Methods
Plant materials
Plant materials were collected from various areas of China. JQK and YGF come from Hangzhou, Zhejiang Province. HQK come from Jiaxing, Zhejiang Province. DJWJ come from Weifang, Shandong Province. KLB and TWWF come from Wuxi Jiangsu Province. MRZ come from Cangzhou Hebei Province.
Genomic DNA extraction
The leaves were collected from the 34 okra accessions. Genomic DNA was extracted from 0.1 g fresh leaves following the improved CTAB protocol [14] and the DNA was detected with 2% agarose gel electrophoresis.
ISSR PCR amplification
DNA primers used in PCR amplification were presented in Table 1. 10 ng template DNA, 10 mL 2x PCR Master Mix, 2x PCR Buffer, 2x dNTP, and 1 mL of each primer were mixed using ultrapure water. All the oligonucleotides were dissolved in Amplification. PCR was performed using ABI 2720 Thermal Cycler (Applied Biosystems, USA) with the following process: 5 minutes pre-denaturation at 94°C, 50 cycles of denaturation at 94°C for 30s, 1 minute for annealing and 72°C extended for 90s. The PCR products were electrophoresed in a 100V with 2% agarose gel for 1 hour and then photographed in a gel imager.
| Primer | Sequence 5'-3' | Tm (℃) | TB | PB | PPB(%) |
|---|---|---|---|---|---|
| 811 | GAGAGAGAGAGAGAGAC | 47 | 9 | 6 | 0.666666667 |
| 823 | TCTCTCTCTCTCTCTCC | 49 | 6 | 4 | 0.666666667 |
| 825 | ACACACACACACACACT | 45 | 9 | 7 | 0.777777778 |
| 830 | TGTGTGTGTGTGTGTGG | 48 | 8 | 7 | 0.875 |
| 834 | AGAGAGAGAGAGAGAGCT | 50 | 8 | 6 | 0.75 |
| 835 | AGAGAGAGAGAGAGAGCC | 54 | 8 | 6 | 0.75 |
| 840 | GAGAGAGAGAGAGAGACT | 49 | 13 | 12 | 0.923076923 |
| 841 | GAGAGAGAGAGAGAGACC | 50 | 12 | 10 | 0.833333333 |
| 842 | GAGAGAGAGAGAGAGATG | 49 | 7 | 5 | 0.714285714 |
| 855 | ACACACACACACACACCT | 52 | 7 | 5 | 0.714285714 |
| 857 | ACACACACACACACACCG | 49 | 6 | 5 | 0.833333333 |
| 884 | ACTAGAGAGAGAGAGAG | 48 | 10 | 3 | 0.3 |
| 887 | GCATCTCTCTCTCTCTC | 49 | 11 | 6 | 0.545454545 |
| 889 | ACTACACACACACACAC | 49 | 7 | 4 | 0.571428571 |
| 891 | ACTTGTGTGTGTGTGTG | 47 | 7 | 4 | 0.571428571 |
Table 1 The annealing temperature (Tm) and the total number of bands (TB), number of polymorphic bands (PB) and % of polymorphic bands (PPB) used in the ISSR primers.
Data statistics
ISSR products were established with 0 and 1. The UPGMA method was used to construct the molecular phylogenetic tree among the accessions via the R program software. PowerMarker software was used to calculate the polymorphism information content (PIC), allele frequency, genetic diversity and genetic distance of each primer. The principal co-ordination analysis (PCoA) of CENTER and EIGEN were obtained in NTSYS-pc [15]. The cluster silhouette plot and optimal number of clusters were obtained by the R program. The two-dimensional array with coordinates was used to identify the 34-point accession differences of okra.
Results
ISSR polymorphism
The 15 ISSR primers were used to amplify 34 okra accessions. A total of 128 DNA fragments was obtained from the 15 primers, and the average fragments amplified from each primer is 8.5, in which polymorphic fragments were 90, accounting for 70.3%. The size of amplified fragments was between 200 and 3000 bp. Different primer has the different amplified results, the primer 840 produced most amplified DNA fragments, as shown in Figure 1. As compared with 840, 830 had the least number of 3 bands with no polymorphism bands. In these 15 primers, there were 7 (GA) n, indicating that there are a lot of (GA) dinucleotide repeats in okra genome.
Figure 1: Agarose gel electrophoresis profiles were obtained by using 840 ISSR primers for the 34 okra accessions. Lanes 1-34 are MRZ-1, MRZ-2, MRZ-3, MRZ-4, MRZ-5, TWWF-1, TWWF-2, TWWF-3, TWWF-4, TWWF-5, KLB-1, KLB-2, KLB-3, KLB-4, KLB-5, DJWJ-1, DJWJ-2, DJWJ-3, DJWJ-4, HQK-1, HQK-2, HQK-3, HQK-4, HQK-5, YGF-1, YGF-2, YGF-3, YGF-4, YGF-5, JQK-1, JQK-2, JQK-3, JQK-4, JQK-5, respectively.
Cluster analysis
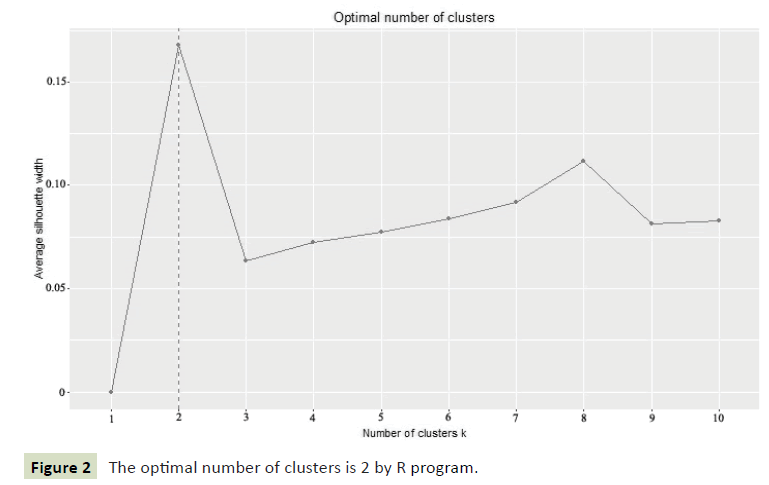
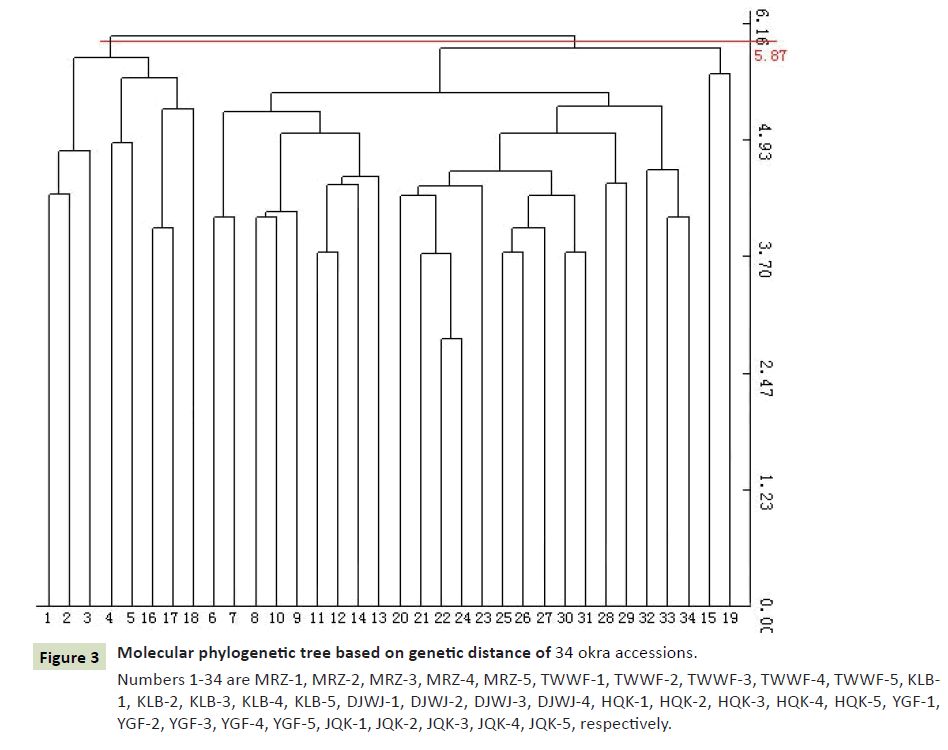
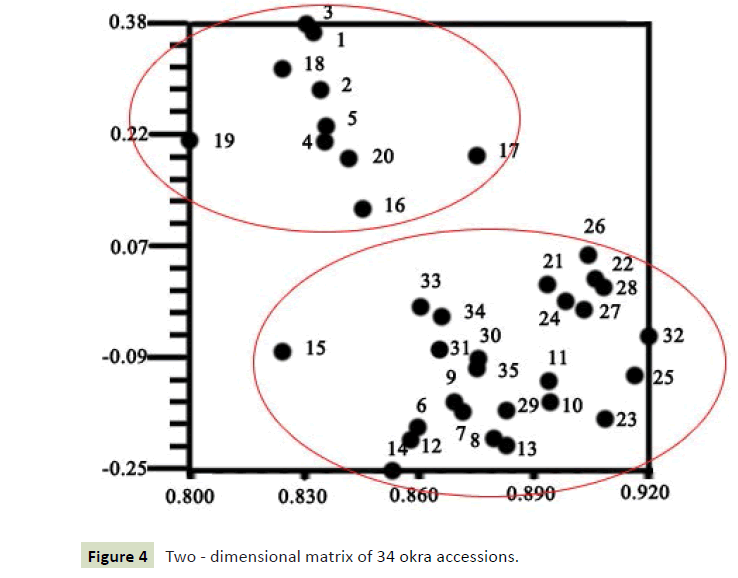
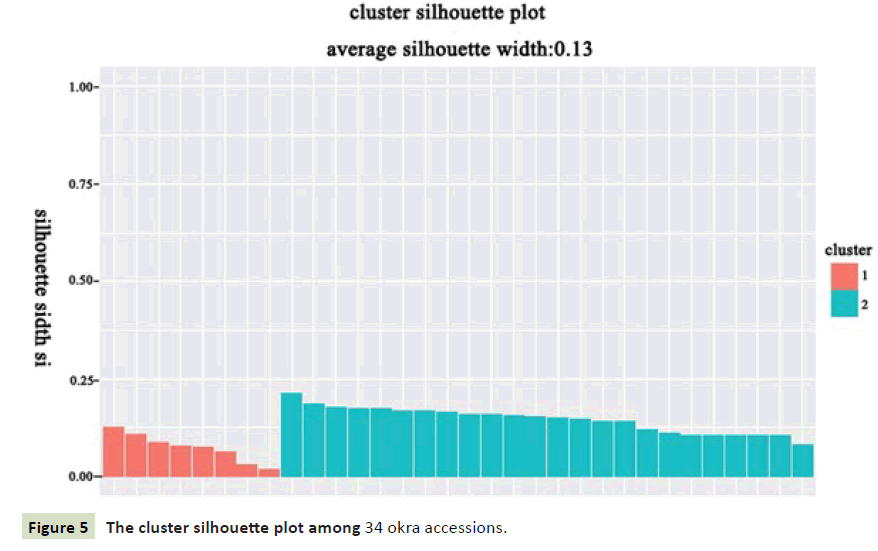
We classify 34 samples against the R program, and the results showed that the two categories were most appropriate (Figure 2). It can be seen from the dendrogram that all materials can be divided into two groups at the threshold of 5.87 (Figure 3). The populations MRZ1-5 and DJWJ1-4 constitute the first group. The second group consists of TWWF1-5, KLB1-5, HQK1-5, YGF1-5 and JQK1-5. The above clustering results are consistent with its geographical distribution. The results from PCoA revealed the diversity among these accessions (Figure 4). The ordination plot indicated clearly distinct groupings and confirmed consistency with those obtained from UPGMA cluster analysis. At the same time, we used the R program to further verify the results and got the cluster silhouette plot (Figure 5). When the width was close to -1, the results of cluster analysis was wrong. When the width was close to +1, the results of cluster was more real. The results obtained were 0.13 indicating that it was credible to classify these 34 accessions into two categories.
Figure 3: Molecular phylogenetic tree based on genetic distance of 34 okra accessions. Numbers 1-34 are MRZ-1, MRZ-2, MRZ-3, MRZ-4, MRZ-5, TWWF-1, TWWF-2, TWWF-3, TWWF-4, TWWF-5, KLB- 1, KLB-2, KLB-3, KLB-4, KLB-5, DJWJ-1, DJWJ-2, DJWJ-3, DJWJ-4, HQK-1, HQK-2, HQK-3, HQK-4, HQK-5, YGF-1, YGF-2, YGF-3, YGF-4, YGF-5, JQK-1, JQK-2, JQK-3, JQK-4, JQK-5, respectively.
The Power Marker software was used to calculate PIC values (Table 2). The results showed that the allele frequency was about 0.5, the genetic diversity was from 0.6857 to 1, the minimum and maximum values of polymorphism were 0.4507 and 0.6801, respectively, and the value of primer 834 was the highest. Based on the 128 DNA fragments generated by ISSR amplification, the genetic similarity matrix of 34 okra accessions was obtained (Table 3). The genetic similarity coefficient (0.94) between HQK- 3 and HQK-5 was the highest, which indicated that the genetic similarity between HQK-3 and HQK-5 was the closest. The similarity coefficient between MRZ-3 and KLB-4 (0.6) was the smallest. The results indicated the genetic similarity between MRZ-3 and KLB-4 was the closest, which was consistent with the result of cluster analysis.
| Primer | Major.Allele. Frquency |
Genotype No | Allele No | Gene Diversity | Heterozygosity | PIC |
|---|---|---|---|---|---|---|
| 811 | 0.5000 | 4.0000 | 5.0000 | 0.6600 | 1.0000 | 0.6115 |
| 823 | 0.5143 | 6.0000 | 6.0000 | 0.6776 | 0.9714 | 0.6447 |
| 825 | 0.5000 | 5.0000 | 6.0000 | 0.6763 | 1.0000 | 0.6370 |
| 830 | 0.5000 | 6.0000 | 7.0000 | 0.6861 | 1.0000 | 0.6519 |
| 834 | 0.5000 | 6.0000 | 7.0000 | 0.7045 | 1.0000 | 0.6801 |
| 835 | 0.5000 | 6.0000 | 7.0000 | 0.6669 | 1.0000 | 0.6224 |
| 840 | 0.5000 | 8.0000 | 9.0000 | 0.6886 | 1.0000 | 0.6556 |
| 841 | 0.5000 | 8.0000 | 9.0000 | 0.6996 | 1.0000 | 0.6728 |
| 842 | 0.5000 | 5.0000 | 6.0000 | 0.6653 | 1.0000 | 0.6191 |
| 855 | 0.5143 | 5.0000 | 5.0000 | 0.6633 | 0.9714 | 0.6219 |
| 857 | 0.5429 | 6.0000 | 6.0000 | 0.6445 | 0.9143 | 0.6064 |
| 884 | 0.6571 | 4.0000 | 4.0000 | 0.5057 | 0.6857 | 0.4507 |
| 887 | 0.5000 | 6.0000 | 7.0000 | 0.6706 | 1.0000 | 0.6283 |
| 889 | 0.5000 | 3.0000 | 4.0000 | 0.6057 | 1.0000 | 0.5293 |
| 891 | 0.5143 | 6.0000 | 6.0000 | 0.6718 | 0.9714 | 0.6357 |
| Mean | 0.5464 | 5.3125 | 5.9375 | 0.6179 | 0.9071 | 0.5792 |
Table 2 The allele frequencies, genetic diversity, heterozygosity and polymorphism information content (PIC) of 15 primers.
| MRZ-1 | MRZ-2 | MRZ-3 | MRZ-4 | MRZ-5 | TWWF-1 | TWWF-2 | TWWF-3 | TWWF-4 | TWWF-5 | KLB-1 | KLB-2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MRZ-1 | 1 | |||||||||||
| MRZ-2 | 0.85 | 1 | ||||||||||
| MRZ-3 | 0.85 | 0.78 | 1 | |||||||||
| MRZ-4 | 0.72 | 0.76 | 0.78 | 1 | ||||||||
| MRZ-5 | 0.75 | 0.74 | 0.76 | 0.81 | 1 | |||||||
| TWWF-1 | 0.66 | 0.67 | 0.67 | 0.71 | 0.69 | 1 | ||||||
| TWWF-2 | 0.65 | 0.66 | 0.67 | 0.7 | 0.67 | 0.87 | 1 | |||||
| TWWF-3 | 0.67 | 0.69 | 0.69 | 0.73 | 0.73 | 0.78 | 0.8 | 1 | ||||
| TWWF-4 | 0.65 | 0.64 | 0.71 | 0.7 | 0.65 | 0.76 | 0.77 | 0.86 | 1 | |||
| TWWF-5 | 0.71 | 0.7 | 0.74 | 0.71 | 0.72 | 0.84 | 0.82 | 0.87 | 0.87 | 1 | ||
| KLB-1 | 0.74 | 0.71 | 0.71 | 0.7 | 0.74 | 0.76 | 0.77 | 0.81 | 0.81 | 0.84 | 1 | |
| KLB-2 | 0.71 | 0.71 | 0.67 | 0.7 | 0.71 | 0.79 | 0.78 | 0.8 | 0.78 | 0.82 | 0.89 | 1 |
| KLB-3 | 0.63 | 0.73 | 0.65 | 0.74 | 0.71 | 0.81 | 0.78 | 0.84 | 0.82 | 0.8 | 0.82 | 0.85 |
| KLB-4 | 0.64 | 0.65 | 0.6 | 0.66 | 0.66 | 0.78 | 0.78 | 0.79 | 0.74 | 0.8 | 0.82 | 0.87 |
| KLB-5 | 0.65 | 0.67 | 0.6 | 0.68 | 0.64 | 0.71 | 0.72 | 0.69 | 0.72 | 0.7 | 0.78 | 0.72 |
| DJWJ-1 | 0.67 | 0.75 | 0.71 | 0.74 | 0.78 | 0.67 | 0.71 | 0.74 | 0.71 | 0.71 | 0.74 | 0.71 |
| DJWJ-2 | 0.74 | 0.74 | 0.78 | 0.73 | 0.81 | 0.7 | 0.72 | 0.74 | 0.75 | 0.78 | 0.75 | 0.77 |
| DJWJ-3 | 0.71 | 0.66 | 0.78 | 0.74 | 0.74 | 0.65 | 0.67 | 0.61 | 0.69 | 0.68 | 0.69 | 0.69 |
| DJWJ-4 | 0.73 | 0.72 | 0.71 | 0.67 | 0.65 | 0.67 | 0.64 | 0.64 | 0.74 | 0.7 | 0.71 | 0.71 |
| DJWJ-5 | 0.76 | 0.69 | 0.75 | 0.73 | 0.67 | 0.7 | 0.69 | 0.72 | 0.72 | 0.74 | 0.75 | 0.75 |
| HQK-1 | 0.78 | 0.74 | 0.71 | 0.69 | 0.75 | 0.74 | 0.78 | 0.74 | 0.73 | 0.74 | 0.81 | 0.74 |
| HQK-2 | 0.76 | 0.77 | 0.75 | 0.76 | 0.78 | 0.78 | 0.78 | 0.77 | 0.74 | 0.79 | 0.78 | 0.77 |
Table 3a Genetic similarity coefficient matrix of 34 okra accessions.
| MRZ-1 | MRZ-2 | MRZ-3 | MRZ-4 | MRZ-5 | TWWF-1 | TWWF-2 | TWWF-3 | TWWF-4 | TWWF-5 | KLB-1 | KLB-2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HQK-3 | 0.69 | 0.71 | 0.7 | 0.74 | 0.75 | 0.81 | 0.79 | 0.79 | 0.76 | 0.81 | 0.84 | 0.85 |
| HQK-4 | 0.77 | 0.76 | 0.71 | 0.71 | 0.74 | 0.72 | 0.73 | 0.76 | 0.78 | 0.8 | 0.84 | 0.82 |
| HQK-5 | 0.71 | 0.71 | 0.71 | 0.74 | 0.75 | 0.78 | 0.79 | 0.79 | 0.78 | 0.81 | 0.84 | 0.82 |
| YGF-1 | 0.74 | 0.74 | 0.76 | 0.77 | 0.75 | 0.77 | 0.76 | 0.76 | 0.79 | 0.81 | 0.79 | 0.76 |
| YGF-2 | 0.72 | 0.73 | 0.73 | 0.74 | 0.71 | 0.78 | 0.78 | 0.79 | 0.78 | 0.8 | 0.76 | 0.76 |
| YGF-3 | 0.76 | 0.78 | 0.77 | 0.73 | 0.74 | 0.76 | 0.77 | 0.81 | 0.77 | 0.78 | 0.8 | 0.77 |
| YGF-4 | 0.65 | 0.67 | 0.66 | 0.71 | 0.7 | 0.76 | 0.74 | 0.8 | 0.77 | 0.76 | 0.77 | 0.77 |
| YGF-5 | 0.7 | 0.71 | 0.69 | 0.71 | 0.7 | 0.73 | 0.71 | 0.78 | 0.75 | 0.79 | 0.77 | 0.74 |
| JQK-1 | 0.78 | 0.77 | 0.75 | 0.73 | 0.71 | 0.76 | 0.78 | 0.78 | 0.77 | 0.79 | 0.8 | 0.77 |
| JQK-2 | 0.74 | 0.78 | 0.74 | 0.76 | 0.71 | 0.78 | 0.8 | 0.81 | 0.8 | 0.81 | 0.81 | 0.8 |
| JQK-3 | 0.73 | 0.66 | 0.72 | 0.7 | 0.71 | 0.71 | 0.74 | 0.74 | 0.74 | 0.74 | 0.74 | 0.75 |
| JQK-4 | 0.71 | 0.72 | 0.69 | 0.7 | 0.71 | 0.73 | 0.72 | 0.75 | 0.77 | 0.76 | 0.74 | 0.74 |
| JQK-5 | 0.67 | 0.68 | 0.68 | 0.69 | 0.71 | 0.74 | 0.74 | 0.79 | 0.79 | 0.81 | 0.74 | 0.74 |
Table 3b Genetic similarity coefficient matrix of 34 okra accessions.
| KLB-3 | KLB-4 | KLB-5 | DJWJ-1 | DJWJ-2 | DJWJ-3 | DJWJ-4 | DJWJ-5 | HQK-1 | HQK-2 | HQK-3 | HQK-4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KLB-3 | 1 | |||||||||||
| KLB-4 | 0.84 | 1 | ||||||||||
| KLB-5 | 0.73 | 0.73 | 1 | |||||||||
| DJWJ-1 | 0.73 | 0.7 | 0.67 | 1 | ||||||||
| DJWJ-2 | 0.73 | 0.73 | 0.67 | 0.88 | 1 | |||||||
| DJWJ-3 | 0.67 | 0.65 | 0.67 | 0.74 | 0.83 | 1 | ||||||
| DJWJ-4 | 0.73 | 0.68 | 0.71 | 0.63 | 0.72 | 0.75 | 1 | |||||
| DJWJ-5 | 0.71 | 0.7 | 0.71 | 0.66 | 0.74 | 0.8 | 0.83 | 1 | ||||
| HQK-1 | 0.74 | 0.77 | 0.79 | 0.76 | 0.79 | 0.71 | 0.67 | 0.71 | 1 | |||
| HQK-2 | 0.78 | 0.74 | 0.69 | 0.78 | 0.78 | 0.74 | 0.69 | 0.72 | 0.87 | 1 | ||
| HQK-3 | 0.83 | 0.81 | 0.73 | 0.74 | 0.76 | 0.7 | 0.67 | 0.68 | 0.83 | 0.88 | 1 | |
| HQK-4 | 0.8 | 0.77 | 0.76 | 0.74 | 0.79 | 0.71 | 0.71 | 0.74 | 0.84 | 0.84 | 0.84 | 1 |
| HQK-5 | 0.81 | 0.8 | 0.73 | 0.78 | 0.79 | 0.73 | 0.67 | 0.73 | 0.86 | 0.9 | 0.94 | 0.86 |
| YGF-1 | 0.78 | 0.74 | 0.73 | 0.76 | 0.79 | 0.79 | 0.73 | 0.78 | 0.81 | 0.84 | 0.8 | 0.81 |
| YGF-2 | 0.78 | 0.75 | 0.73 | 0.73 | 0.78 | 0.74 | 0.73 | 0.78 | 0.84 | 0.85 | 0.83 | 0.8 |
| YGF-3 | 0.81 | 0.73 | 0.71 | 0.75 | 0.77 | 0.72 | 0.69 | 0.77 | 0.85 | 0.86 | 0.82 | 0.78 |
| YGF-4 | 0.81 | 0.76 | 0.75 | 0.74 | 0.72 | 0.67 | 0.64 | 0.71 | 0.76 | 0.77 | 0.82 | 0.74 |
| YGF-5 | 0.78 | 0.73 | 0.71 | 0.74 | 0.72 | 0.69 | 0.61 | 0.71 | 0.78 | 0.81 | 0.82 | 0.78 |
| JQK-1 | 0.76 | 0.74 | 0.75 | 0.75 | 0.78 | 0.74 | 0.67 | 0.77 | 0.87 | 0.81 | 0.82 | 0.85 |
| JQK-2 | 0.82 | 0.74 | 0.75 | 0.78 | 0.78 | 0.71 | 0.69 | 0.74 | 0.84 | 0.86 | 0.85 | 0.84 |
| JQK-3 | 0.71 | 0.71 | 0.71 | 0.67 | 0.74 | 0.72 | 0.63 | 0.67 | 0.78 | 0.77 | 0.79 | 0.79 |
| JQK-4 | 0.73 | 0.7 | 0.74 | 0.74 | 0.74 | 0.69 | 0.67 | 0.67 | 0.78 | 0.8 | 0.79 | 0.76 |
| JQK-5 | 0.74 | 0.72 | 0.73 | 0.74 | 0.78 | 0.7 | 0.65 | 0.71 | 0.77 | 0.79 | 0.83 | 0.78 |
Table 3c Genetic similarity coefficient matrix of 34 okra accessions.
| HQK-5 | JQK-1 | JQK-2 | JQK-3 | JQK-4 | JQK-5 | |
|---|---|---|---|---|---|---|
| HQK-5 | 1 | |||||
| YGF-1 | 0.83 | |||||
| YGF-2 | 0.83 | |||||
| YGF-3 | 0.84 | |||||
| YGF-4 | 0.81 | |||||
| YGF-5 | 0.84 | |||||
| JQK-1 | 0.87 | 1 | ||||
| JQK-2 | 0.85 | 0.89 | 1 | |||
| JQK-3 | 0.78 | 0.83 | 0.84 | 1 | ||
| JQK-4 | 0.76 | 0.78 | 0.84 | 0.86 | 1 | |
| JQK-5 | 0.81 | 0.82 | 0.84 | 0.81 | 0.87 | 1 |
Table 3d Genetic similarity coefficient matrix of 34 okra accessions.
Discussion
Microsatellite markers, a significant tools for genetic analyses, can be distinguished with a high degree of variability. However, the common protocols used for separating Microsatellites suffer from cost, time consuming, laborious [16,17] and inefficiency, which limit their applications. To overcome the limitation and further improve yield, ISSR-PCR were introduced. Furthermore, in genome only the regions that are abundant in Microsatellite motifs are targets for ISSR. The structure of Microsatellite was efficient for the strategy of ISSR-PCR with anchored 3’ and 5' terminal in the application of generating polymorphic loci among different species, which are certified succeed due to more than 86% consisted of internal microsatellite sequence among all the products except these at terminals. Agarose gel electrophoresis is the most common approach for isolating PCR products, which can achieve polymorphic bands with lower percentage, but have the advantages of cheap and time saving. Therefore, in this study we choose agarose gel for separating different bands generating in ISSR-PCR.
Molecular basis of polymorphism and their distribution across the genome is quite important for study on genetic diversity and population structure. In this study, the genetic diversity among 34 accessions of okra was analyzed using ISSR markers. A total of 128 DNA fragments and 90 polymorphic fragments (70.3%) were obtained by PCR amplification from 34 accessions, indicating that the genetic diversity of okra was relatively abundant. These fragments were processed to construct the molecular phylogenetic tree. The accessions of okras were divided into two major groups at the threshold of 5.87, which was consistent with the results obtained by the R program. Meanwhile, from the twodimensional matrix obtained by NTSYS-PC software, the 34 okras were more appropriately divided into two groups. The above results showed that the two taxa were reliable.
From the constructed molecular dendrogram and the genetic similarity coefficient matrix, it can be seen that the HQK-3 and HQK-5 accessions had the closest genetic relationship, while the MRZ-3 and KLB-4 had the furthest genetic relationship. The phylogenetic relationships of different accessions are sometimes not obvious, but the genetic relationship of the same accessions is very different, which indicated that the genetic relationship between okra germplasms was complex.
Information on genetic diversity and population structure is essential for a better understanding of the breeding and genetic relationships of crop germplasm. The values of allele frequency, genetic diversity, and heterozygosity were high, which showed that the ISSR primers can produce some measurable fragments. PIC provided an estimate of the discriminant ability of markers based on marker NA and allele relative frequencies to distinguish genotypes [18] PIC was used to determine the genetic diversity of okra. High, moderate and low polymorphisms were expressed as PIC> 0.5, 0.5> PIC> 0.25 and PIC <0.25, respectively [19,20]. The PIC value ranged from 0.4507 to 0.6801, with an average value of 0.5745 in this study, indicating that the primers could form high polymorphisms, which were useful for genetic variation in the genotype studied in this study. The above results confirm the potential value of genetic diversity preservation for future breeding of Okra.
Conclusion
The phenotypic and genotypic variations are important for the development of new improved lines and cultivars. It would be a good tool of selecting okra breeding program to study the characterization of the genetic variability among okra germplasm. In the present study, the considerable genetic diversity among 34 okra accessions was observed at both the morphological and molecular levels, which is of importance for germplasm classification, management, and further utilization. The results of this paper showed the ISSR markers would be a useful tool for okra genetic diversity studies and in breeding programs. The phylogenetic trees based on genetic distance indicated the 34 accessions were divided into two major groups. Moreover, the two-dimensional matrix also confirmed clustering results. On the whole, the data showed that there was a high degree of genetic diversity among the okra accessions revealed by the ISSR markers. Our results confirm the potential value of genetic diversity preservation for future breeding of Okra.
Conflict of Interest
The author confirms that this article content has no conflict of interest.
Acknowledgments
This research was supported by grants from Key Research & Development Project of Hunan Provincial Department of Science and Technology (2016WK2003), National Key Research and Development Program (2017YFF0210301), National Key Laboratory of Plant Molecular Genetics (2015), National Natural Science Foundation of China (31540064, 31071076 and 30871325), Program for New Century Excellent Talents in University (NCET-10-0363), Excellent Youth Foundation of Hunan Province (11JJ1005), Ph.D. Programs Foundation of Ministry of Education of China (20130161110005), and Hunan Provincial Innovation Foundation for Postgraduate (CX2015B073, CX2016B097, CX2017B to Cheng Zhang).
References
- Kochhar SL (1986) Tropical Crops. Macmillan Publishers Ltd. London and Basigstoke pp 467.
- Schafleitner R, Kumar S, Lin CY, Hegde SG, Ebert A (2013) The Okra (Abelmoschus esculentus) transcriptome as a source for gene sequence information and molecular markers for diversity analysis. Gene 517 : 27-36.
- Çalışır S, Özcan M, Hacıseferoğulları H, Yıldız MU (2005) A study on some physico-chemical properties of Turkey Okra ( Hibiscus esculenta, L.) seeds. Food Eng 68 : 73-78.
- Oyelade OJ, Ade-Omowaye BIO, Adeomi VF (2003) Influence of variety on protein, fat contents and some physical characteristics of Okra seeds. Food Eng 57 : 111-114.
- Gul MZ, Bhakshu LM, Ahmad F, Kondapi AK, Qureshi IA, et al. (2011) Evaluation of Abelmoschus moschatus extracts for antioxidant, free radical scavenging, antimicrobial and antiproliferative activities using in vitro assays. BMC Complement Altern Med 11: 1-12.
- Yuan CY, Zhang C, Wang P, Hu S, Chang HP, et al. (2014) Genetic diversity analysis of Okra (Abelmoschus esculentus L.) by Inter-simple Sequence Repeat (ISSR) markers. Genet Mol Res 13: 3165-3175.
- Zhang XY, Huang J, Liu GD (2009) ISSR Analysis of 43 Okra Germplasms. Chin J Trop Crops 3: 293-298.
- Huang J, Chen XB, Ye HL, Liu GD (2008) ISSR Analysis of 16 Okra germplasms. Chinese Agricultural Science Bulletin 5: 403-408.
- Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by Simple Sequence Repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20: 176-183.
- Wu YT, Zhang TZ, Yin JM (2001) Genetic diversity of upland Cotton accessions detected by molecular marker and morphological traits. J Genet Genomics 28:1040-1050.
- Ghariani S, Trifi-Farah N, Chakroun M, Marghalin S, Marrakchi M (2003) Genetic diversity in Tunisian perennial ryegrass revealed by ISSR markers. Genetic Gen Resour Crop Evol 50: 809-815.
- Huang JC, Sun M (2000) Genetic diversity and relationships of sweet potato and its wild relatives in Jpomoea series Batatas (Convolvlaceae) as revealed by ISSR and restriction analysis of chloroplast DNA.Theor Appl Genet 100 : 1050-1060.
- Shuldiner AR, Nirula A, Roth J (1990) RNA template-specific polymerase chain reaction (RS-PCR): A novel strategy to reduce dramatically false positives. Gene 91:139-142.
- Doyle J (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13-15.
- Rohlf FJ (2009) NTSYSpc: Numerical taxonomy system, ver. 2.21c. Exeter Software, Setauket.
- Sethy NK, Shokeen B, Edwards KJ, Bhatia S (2006) Development of microsatellite markers and analysis of intraspecifc genetic variability in Chickpea (Cicer arietinum L.). Theor Appl Genet 112: 1416-1428.
- Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: A review. Mol Ecol 11: 1-16.
- Nagl N, Taski-Ajdukovic K, Popovic A, Curcic A (2011) Estimation of genetic variation among related sugar beet genotypes by using RAPD. Genetika 43 : 575-582.
- Xie W, Zhang X, Cai H, Liu W (2010) Genetic diversity analysis and transferability of cereal EST-SSR markers to Orchardgrass (Dactylis glomerata L.). Biochem Syst Ecol 38: 740-749.
- Vaiman D, Mercier D, Moazami-Goudarzi K, Eggen A, Ciampolini R et al. (1994) A set of 99 cattle Microsatellites: characterization, synteny mapping, and polymorphism. Mamm Genome 5: 288-297.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences