Expression of Adipocyte Fatty Acid-Binding Protein Gene in Abdominal Adipose Tissue and Its Association with Growth and Fatness Traits in Commercial Meat Type Chickens
1Department of Animal Production, Faculty of Agriculture, Cairo University, Giza, Egypt
2Department of Animal and Avian Sciences, University of Maryland, College Park, USA
- *Corresponding Author:
- Mona M Ghaly
Department of Animal Production, Faculty of Agriculture,
Cairo University, Gamma St.,
Giza, Egypt
Tel: 00201002828243
E-mail: Monaghaly19@yahoo.com
Received date: August 03, 2018; Accepted date: August 09, 2018; Published date: August 17, 2018
Citation: Ghaly MM, Li M (2018) Expression of Adipocyte Fatty Acid-Binding Protein Gene in Abdominal Adipose Tissue and Its Association with Growth and Fatness Traits in Commercial Meat Type Chickens. J Genom Gene Study Vol.1 No.1:5
Abstract
Adipocyte fatty acid-binding protein (A-FABP) gene expression was assessed in abdominal fat tissues of five commercial meat-type chicken hybrids (Aviagen, Arbor Acre, Hubbard, Cobb and Ross) at of 37 days old. Real-time quantitative reverse transcription polymerase-chain reaction was used. The relative A-FABP gene mRNA expression level was calculated with 2-ΔΔCt method using males as calibrators for their target females. The meat type hybrids were diverse in their genetic makeup and response. Aviagen females recorded significant 2.176 more than their males. Hubbard females recorded non-significant 0.6533 fold, same as their males. Arbor acres females are lower than their males by significant 0.1243 fold. Both Cobb and Ross genotypes scored significant 0.3723 and 0.3951, respectively, fold than their males. Hierarchical clustering analysis dendograme method merged Cobb and Ross genotypes to the first closest cluster. Both Arbor Acres and Hubbard had joined into a further cluster. Aviagen genotype was much closer to Arbor Acres and Hubbard nest. Lower abundance of A-FABP gene expression for Arbor Acres was significantly associated with growth and most of carcass parameters retardation. Lower expression of A-FABP gene for Cobb genotype had a unique elevation response for both growth and most of carcass parameters and strong positive association with abdominal fat deposition, especially for males. So, the A-FABP gene could be linked to major gene(s) that influence the abdominal fat content in case of Cobb broilers. FABP4 may provide useful information for further studies on its roles in growth, carcass traits and fatness for both Arbor Acres and Cobb hybrids.
Keywords
Gene expression; A-FABP gene; Growth performance; Fat deposition; Meat type chicken
Introduction
Excessive adiposity is a problem in modern fast growing broiler industry. Selection in broiler chickens for increased growth rate has resulted in higher body fat deposition, which is considered as a by-product with very low commercial benefit [1]. Although several strategies of selection for leanness in poultry production have been defined, measuring of body fat content, such as abdominal fat (AF), skin fat (SF) and intramuscular fat (IMF), as a major determinant of chicken meat quality, is still difficult because of its tediousness and expensiveness [2,3]. Knowing the molecular mechanisms of growth will add to a more efficient selection process for growth in broiler chickens. Basically, there are two major methods of quantitative trait loci (QTL) determination, the candidate gene approach and the whole-genome scanning [4]. The candidate gene approach is used to detect QTL responsible for genetic variation in the traits of interest. Researchers and producers have paid attention and found several candidate genes or markers for chicken fat traits [5-7].
Fatty acid-binding proteins (FABPs) are a group of carrier-proteins for fatty acids and lipophilic substances like eicosanoids and retinoids [8,9]. These proteins can be divided into two main groups; one was associated with the plasma membrane (F-ABPPM) and the other with the intracellular or cytoplasmic proteins (F-ABPC) [10]. FABPs can reversibly bind to lipids, hydrophobic molecules such as saturated and unsaturated long-chain fatty acids (FAs) and eicosanoids, with high affinity and selectivity [11]. The overall FABP gene structure is conservative in all family members and consists of four exons separated by three introns [12]. The exon/intron sites are similar, but the length of intron is changeable among the genes [11,13,14]. FABPs have actively facilitated the transport of FAs to the cell for lipid oxidation in the mitochondrion, regulation of lipid-mediated transcription in the nucleus, trafficking, signaling, membrane construction in the endoplasmic reticulum, and regulation of enzyme activity and storage as lipid droplets in the cytoplasm [14]. FABP gene is an important factor that controls intramuscular fat content, which in turn controls meat tenderness and flavor [15]. FABP gene has been shown to be associated with lipid metabolism (lipolysis and lipogenesis), homeostasis in adipocytes, marbling and back fat deposition [9,16]. The FABP4 gene is found to be significantly related to meat tenderness in sheep [17]. The A-FABP was selected as candidate gene for regulating intramuscular fat metabolism in pigs [18] and Carcass weight and marbling [19]. In chicken, the A-FABP gene was used as a marker to identify intramuscular fat acceleration [20]. In cattle, genetic polymorphisms of FABP4 gene were found to be related to meat quality grades [21,22]. In milk, Nafikov et al. [23] showed that some FABP4 haplotypes are correlated with specific fatty acid characters, regardless to differences in milk yield. Also, Liu ZW et al. [24] declared that the overexpression of cattle A-FABP gene in transgenic mice resulted in remarkable increase in TG content. A-FABP promotes the conversion of T4 to T3 in brown adipocytes which increases thermogenesis. In addition, thermo genic stimuli in mice were accompanied by increased levels of A-FABP in both white and brown adipose tissues and the bloodstream [25]. The main aims of this study were to detect the relative expression of A-FABP gene in different commercial meat-type chickens using FQ-RT-PCR and its associations with growth, body composition and fatness traits. It is important to observe the association between transcription levels of the A-FABP and the intramuscular fat (IMF) contents, to provide insights into confirming possible associations and to evaluate the genetic architectures for these genotypes.
Materials and Methods
Experimental birds, diets and tissue sampling
This experiment was carried out at Poultry Research Station, Faculty of Agriculture, Cairo University, Egypt. Five broiler genotypes were used in this study and included the following hybrids: Aviagen, Arbor Acre, Hubbard, Cobb and Ross. A total of 5000 chicks, 1000 per each genotype, were used. The chicks were fed a standard starting diet until they reached 14 days old. Then, they received a standard growing diet until 37 days old. Birds were allocated in equal numbers in floor pens and were maintained with a 16-h light and 8-h dark cycle in a temperature-controlled environment with ad libitum access to feed and drink. In addition, body weights (BW) of different ages were recorded until the age of 37 days. Ten birds were killed by cervical dislocation at 37 days of age (five for each sex). Carcass traits were measured for the same harvested chickens at the age of 37 days, including carcass weight (CW), breast width (BW), fore half (FHW) and dorsal half (DHW) muscle weight, breast major (MPMW) and small (SPMW) pectoral is muscle weight, thigh muscle weight (TMW), drum muscle weight (DMW), shank length (SL), head weight (HW), neck weight (NW), wing weight (WW), all edible parts (heart, liver, spleen and gizzard) weight, and abdominal fat (AF). Also, body measurements relative to the carcass weight were recorded. Growth efficiency (GE) and specific growth rate (SGR) were calculated according to Gondwe and Wollny [26]. Percentage of total muscular fat (TMF) content was determined according to the international organization for standardization ISO (1973). Both carcass traits and edible part traits were expressed as absolute and percentage of carcass weight at slaughter age (37 days).
Gene expression analysis
RNA extraction and reverse transcription-PCR assay for A-FABP gene expression
About 0.5 g tissue from abdominal fat was aseptically removed after slaughter and placed in RNA later solution and kept at -80°C until the time of analysis. Total RNA was isolated from five chicken abdominal fat tissues per sex (5♂ + 5♀) per genotype, using Qiagen’s RNeasy Lipid Tissue Mini Kit and Qiazol lysis reagent procedure according to the manufacturer’s protocol Qiagen, (Germany). The quantity and integrity of isolated RNA were determined for each sample by using NanoDropTM 2000 Spectrophotometer-Thermo Scientific Inc (Wilmington, Delware- USA). RNA samples were stored at -80°C until use. Reverse Transcription (RT) Polymerase Chain Reaction (PCR) was performed using a High Capasity cDNA reverse transcription kit containing RNA (1 μg), 20 pmol gene-specific primer, 9700 GeneAmp PCR-Applied Biosystems (California, USA). The mixture was incubated at 25°C for 10 min for enzyme activation, 37°C for 120 min, 85°C to deactivate the enzyme, and then stored at -20°C. A chicken A-FABP fragment (138 bp) was amplified with a sense primer (5'-AAGACTGCTACCTGGCCTGA-3') and an antisense primer (5'-TCCCTTCCCCAGACACAATA-3'). The primers were designed according to the sequences of A-FABP gene in Gallus gallus, which was used as target gene. Chicken ribosomal 18S RNA was chosen as a reference gene. The fragment size was 148 bp, the sense primer was (5'-CGCGTGCATTTATCAGACCA-3'), and the anti-sense primer was (5'-ACCCGTGGTCACCATGGTA-3'), (Primer- Invetrogen, USA).
Real-time PCR testing on mRNA level in abdominal fat tissue
A-FABP mRNA gene quantitation from abdominal fat tissue was assessed by real-time RT-PCR using a master mix containing SYBRTM Green PCR Master Mix- Life Technologies (California, US). Ten pmol forward primer, 10 pmol reverse primer, cDNA, and water to perform real-time PCR. The following PCR protocol was used on the 500 Real-Time PCR System-Applied BiosystemsTM (California USA). Initial steps include 2 min at 50°C and 10 min at 95°C, followed by two-step amplification program (15 sec at 95°C followed by 1 min at 61°C) and repeated 45 times. Runs were performed in three technical replicates per sample.
Statistical analysis
Expression levels of mRNA, as cycle threshold values, for each gene (A-FABP and 18S) were deviated from its cycle threshold values for ribosomal 18S RNA (housekeeping gene). The relative quantification method was conducted following the equations: ΔCt=CtA-FABP – Ct18S. After all the ΔCt values were obtained for all biological and technical replicates, the mean ΔCt values for each female genotype were compared to the mean ΔCt for its male (calibrator). Thus, all the five genotypes data for the A-FABP gene were expressed as the fold-change relative to male genotype. The amount of target molecules relative to the calibrator males were calculated by 2− ΔΔCT method.
Data was analyzed using SAS 9.1. The model included genotype and sex as main fixed effects; the individual bird was the experimental unit for gene expression analysis. Gene expressionphenotype association analysis and contrast were performed by SAS GLM procedure. The genetic effects were analyzed by fixed procedure according to the following model: Y=μ+G+S+e, where Y=an observation on the trait, μ=the overall population mean, G=the fixed effect of genotype, S=the fixed effect of sex and e=the residual random error. The significant associations were calculated using simple linear regression as the following model: Y=b0+b1X+e where Y=the dependent phenotypic variable, X=the independent target gene expression variable deviated from its housekeeping gene, b0=the intercept and b1=the association of gene effect and e=the residual random error. Clustering procedures used to calculate nearest neighbor hierarchical method by computer program SAS 9.1.
Results
Expression levels of the fat deposition gene among genotype groups
Least squares analysis of variance means
Different comparisons between genotypes had been observed in Tables 1-3. For combined sexes as presented in Table 1, difference in expression level among genotypes showed that Ross genotype recorded the highest ΔCT mean (16.15) (lowest expression) of all. Least squares means of ΔCT for Hubbard group recorded the lowest (highest expression) value (14.03) but is not significantly different from Aviagene genotype. Both sCobb and Arbor Acre recorded intermediate values and are significantly the same. As shown in Table 2, least squares mean of ΔCT for Ross males was significantly the highest of all genotypes (15.74) (the lowest expression). Meanwhile, Hubbard males scored significantly the lowest least squares mean at all (13.89) (the highest expression). Least squares means of females at Table 3. Revealed that Aviagene females scored the lowest least squares mean at all (14.00), (the highest expression) but it does not significantly differ from Hubbard females. Ross females recorded highest least squares mean of all (16.55), (the lowest expression) but not significantly differed from both Cobb and Hubbard females.
Table 1: Least squares means of ΔCT ± standard errors for different genotypes (both sexes) and linear pair genotype contrasts and contrast versus the rest (linear function ± SE).
n: Number of missed replicate; ΔCT1: It represent significance within column with different super alphabetic.
| Versus the rest | p-Value | F-Value | Contrast SS | Estimate | Versus | ∆CT1 | Genotype{5* (3rep-n)} | |||
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | F-Value | Contrast SS | Estimate | |||||||
| 0.8428 | 0.04 | 0.04722718 | 0.044 ± 0.225 | 0.0099 | 6.85 | 8.19146257 | 0.739 ± 0.284 | Arbor Acre | 14.28c ± 0.201 | Aviagen-30 |
| 0.0001 | 15.8 | 18.90277404 | -1.122 ± 0.282 | Hubb | ||||||
| 0.1362 | 2.5 | 2.68780879 | -0.423 ± 0.282 | Cobb | ||||||
| 0.0007 | 12.15 | 14.53388355 | 0.984 ± 0.282 | Ross | ||||||
| 0.0001 | 15.51 | 18.5590087 | -0.879 ± 0.225 | 0.0099 | 6.85 | 8.19146257 | 0.738 ± 0.284 | Aviagen | 15.02b ± 0.201 | Arbor Acre-30 |
| <0001 | 43.44 | 51.98129077 | -1.861 ± 0.284 | Hubb | ||||||
| <0001 | 16.93 | 20.26374474 | -1.162 ± 0.282 | Cobb | ||||||
| 0.3865 | 0.75 | 0.90299399 | 0.245 ± 0.282 | Ross | ||||||
| <.0001 | 42.03 | 50.2920122 | 1.447 ± 0.228 | 0.0001 | 15.8 | 18.90277404 | -1.122 ± 0.283 | Aviagen | 14.03c ± 0.201 | Hub -30 |
| <0001 | 43.44 | 51.98129077 | 1.861 ± 0.283 | Arbor Acre | ||||||
| 0.0145 | 6.13 | 7.33477107 | 0.699 ± 0.282 | Cobb | ||||||
| <0001 | 55.65 | 66.58666313 | 2.106 ± 0.282 | Ross | ||||||
| 0.0113 | 6.6 | 7.89341517 | 0.573 ± 0.228 | 0.1362 | 2.25 | 2.68780879 | -0.423 ± 0.283 | Aviagen | 15.45b ± 0.204 | Cobb-29 |
| <0001 | 16.93 | 20.26374474 | 1.162 ± 0.287 | Arbor Acre | ||||||
| 0.0145 | 6.13 | 7.33477107 | -0.699 ± 0.284 | Hubb | ||||||
| <0001 | 24.84 | 29.72198034 | 1.407 ± 0.282 | Ross | ||||||
| <0001 | 28.21 | 33.7620232 | -1.186 ± 0.225 | 0.0007 | 12.15 | 14.53388355 | 0.984 ± 0.284 | Aviagen | 16.15a ± 204 | Ross-29 |
| 0.3865 | 0.75 | 0.90299399 | -0.245 ± 0.284 | Arbor Acre | ||||||
| <0001 | 55.65 | 66.58666313 | -2.106 ± 0.284 | Hubb | ||||||
| <0001 | 24.84 | 29.72198034 | -1.407 ± 0.282 | Cobb | ||||||
Table 2: Least squares mea of ∆CT ± standard errors for different males’ genotypes and linear pair genotype contrasts and contrasts versus the rest (linear function ± SE).
n: Number of missed replicate; ∆CT1: It represent significance within column with different super alphabetic.
| Versus the rest | p-Value | F-Value | Contrast SS | Estimate | Versus | ∆CT | Genotype {5* (3rep-n)} |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | F-Value | Contrast SS | Estimate | |||||||
| 0.0207 | 5.6 | 3.48103454 | -0.538 ± 0.227 | 0.3115 | 1.04 | 0.64575489 | -0.293 ± 0.287 | Arbor Acre | 14.56bc ± 0.284 | Aviagen-15 |
| <0001 | 26.22 | 16.2912115 | -1.473 ± 0.287 | Hubb | ||||||
| 0.009 | 7.21 | 4.48296292 | -0.773 ± 0.287 | Cobb | ||||||
| 0.1842 | 1.8 | 1.11745545 | 0.385 ± 0.287 | Ross | ||||||
| 0.4528 | 0.57 | 0.35422457 | -0.171 ± 0.227 | 0.3115 | 1.04 | 0.64575489 | -0.293 ± 0.287 | Aviagen | 14.27cd ± 0.284 | Arbor Acre-15 |
| 0.0001 | 16.82 | 10.45001663 | -1.180 ± 0.287 | Hubb | ||||||
| 0.1001 | 2.78 | 1.72583948 | -0.479 ± 0.287 | Cobb | ||||||
| 0.021 | 5.57 | 3.46215392 | 0.679 ± 0.287 | Ross | ||||||
| <0001 | 32.82 | 20.395145 | 1.303 ± 0.227 | <0001 | 26.22 | 16.29121159 | -1.473 ± 0.287 | Aviagen | 13.89d ± 0.284 | Hubb-15 |
| 0.0001 | 16.82 | 10.45001663 | 1.180 ± 0.287 | Arbor Acre | ||||||
| 0.0175 | 5.93 | 3.6823171 | 0.700 ± 0.287 | Cobb | ||||||
| <.0001 | 41.75 | 25.94206101 | 1.859 ± 0.287 | Ross | ||||||
| 0.0643 | 3.53 | 2.19630481 | 0.427 ± 0.227 | 0.009 | 7.21 | 4.48296292 | -0.773 ± 0.287 | Aviagen | 15.04b ± 0.284 | Cobb-15 |
| 0.1001 | 2.78 | 1.72583948 | 0.479 ± 0.287 | Arbor Acre | ||||||
| 0.0175 | 5.93 | 3.6823171 | -0.700 ± 0.287 | Hubb | ||||||
| <.0001 | 16.22 | 10.0768059 | 1.159 ± 0.287 | Ross | ||||||
| <0001 | 20.14 | 12.51158 | -1.021 ± 0.227 | 0.1842 | 1.8 | 1.11745545 | 0.385 ± 0.287 | Aviagen | 15.74a ± 0.284 | Ross-15 |
| 0.021 | 5.57 | 3.46215392 | -0.679 ± 0.287 | Arbor Acre | ||||||
| <.0001 | 41.75 | 25.94206101 | -1.859 ± 0.287 | Hubb | ||||||
| 0.0001 | 16.22 | 10.0768059 | -1.159 ± 0.287 | Cobb | ||||||
Table 3: Least squares mea of ∆CT ± standard errors for different females’ genotypes and linear pair genotype contrasts and contrast versus the rest (linear function ± SE).
n: Number of missed replicate; ∆CT1:It epresents significance within column with different super alphabetic (P<0.05).
| Versus the rest | p-Value | F-Value | Contrast SS | Estimate | Versus | ∆CT | Genotype {5* (3rep-n)} | |||
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | F-Value | Contrast SS | Estimate | |||||||
| 0.1071 | 2.67 | 4.72230866 | 0.627 ± 0.384 | 0.0005 | 13.28 | 23.5338638 | 1.771 ± 0.486 | Arbor Acre | 14.00b ± 0.284 | Aviagen-15 |
| 0.117 | 2.52 | 4.46216758 | -0.771 ± 0.486 | Hubb | ||||||
| 0.8803 | 0.02 | 0.04049631 | -0.073± 0.486 | Cobb | ||||||
| 0.0017 | 10.6 | 18.78663761 | 1.582 ± 0.486 | Ross | ||||||
| <.0001 | 17.06 | 30.2201764 | -1.586 ± 0.384 | 0.0005 | 13.28 | 23.53386384 | 1.771 ± 0.486 | Aviagen | 15.77a ± 0.284 | Arbor Acre-15 |
| <.0001 | 27.37 | 48.49110831 | -2.542 ± 0.486 | Hubb | ||||||
| 0.0003 | 14.41 | 25.52683008 | -1.844 ± 0.486 | Cobb | ||||||
| 0.699 | 0.15 | 0.26709866 | -0.188 ± 0.486 | Ross | ||||||
| 0.0001 | 17.15 | 30.3938333 | 1.591 ± 0.384 | 0.117 | 2.52 | 4.46216758 | -0.771 ± 0.486 | Aviagen | 14.19b ± 0.284 | Hubb-15 |
| <.0001 | 27.37 | 48.49110831 | 2.542 ± 0.486 | Arbor Acre | ||||||
| 0.1555 | 2.06 | 3.6524843 | 0.697 ± 0.486 | Cobb | ||||||
| <.0001 | 23.46 | 41.5604545 | 2.354 ± 0.486 | Ross | ||||||
| 0.0654 | 3.5 | 6.2064308 | 0.719 ± 0.384 | 0.8803 | 0.02 | 0.04049631 | -0.073 ± 0.486 | Aviagen | 15.85a ± 0.294 | Cobb-14 |
| 0.0003 | 14.41 | 25.52683008 | 1.844 ± 0.486 | Arbor Acre | ||||||
| 0.1555 | 2.06 | 3.6524843 | -0.697 ± 0.486 | Hubb | ||||||
| 0.0011 | 11.61 | 20.57159893 | 1.656 ± 0.486 | Ross | ||||||
| 0.0008 | 12.36 | 21.9036208 | -1.351 ± 0.384 | 0.0017 | 10.6 | 18.78663761 | 1.582 ± 0.486 | Aviagen | 16.55a ± 0.294 | Ross-4 |
| 0.699 | 0.15 | 0.26709866 | 0.188 ± 0.486 | Arbor Acre | ||||||
| <.0001 | 23.46 | 41.5604545 | -2.354 ± 0.486 | Hubb | ||||||
| 0.0011 | 11.61 | 20.57159893 | -1.656 ± 0.486 | Cobb | ||||||
Linear contrasts in two-way analysis of variance
Contrasts facilitate comparisons among groups widely and observe difference between specific pairs of groups [27-29]. In general for both sexes, only Aviagene genotype recorded lowest ΔCT least squares means over the rest (+ 0.044) and become not significantly superior over the rest (p 0.8428). In the same time, Results of linear contrasts given in Table 2 for males revealed that ΔCT least squares means for both Arbore Acre and Hubbard genotypes is not significantly superior over the rest. (p=0.4528 and p=0.0643, respectively). Results of linear contrasts for females given in Table 3 revealed that ΔCT least squares means for both Aviagene and Cobb is significantly the same(p=0.8803). They had ΔCT mean 0.627 and 0.719, respectively over the rest and both are not significantly superior over the rest. (p=0.1071 and p=0.0654, respectively) and their least square mean superiority over the rest was extremely the same (p=0.627 and p=0.719 respectively). Arbor Acres and Ross had significantly the same ΔCT but they both had significantly lower ΔCT from the rest (-1.586 and -1.351 respectively).
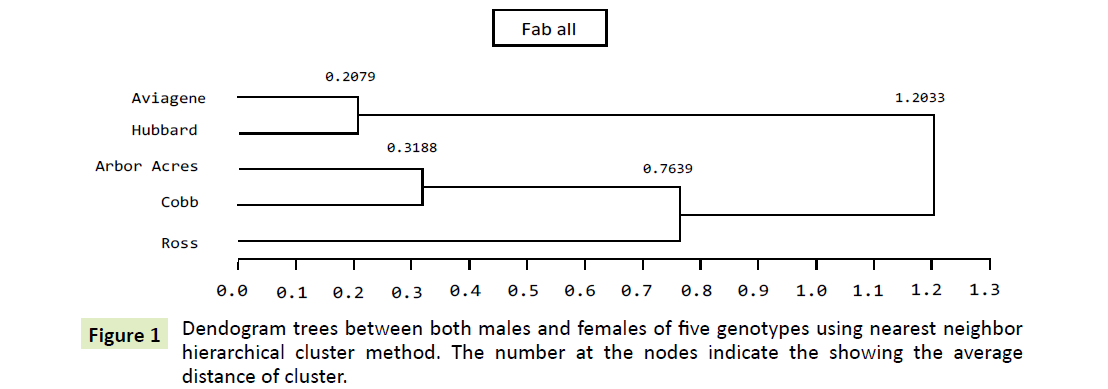
Genotypes allocation to clusters and cluster distance
Combined sexes: As shown in Figures 1 and 2, phylogenetic tree for five independent genotype populations by the nearest neighbor method shows three distinct clusters. The first one aggregates the, Aviagene and Hubbard populations. Their least square means are significantly the same (Table 1), they seems to be the most homologous groups. Their inter cluster distance was lowest among all clusters (0.2079). The second cluster compresses Arbor Acre and Cobb genotypes at 0.3118 point of distance. Similar genotypes possess the existence of high gene flow [30]. The third one, Ross genotype formed its own branch (cluster) to the closest cluster to it, Arbor Acre and Cobb at 0.7639 degree of distance. Finally, the dendogram distances obtained by the nearest-neighbor method for the five genotype populations was (1.2033).
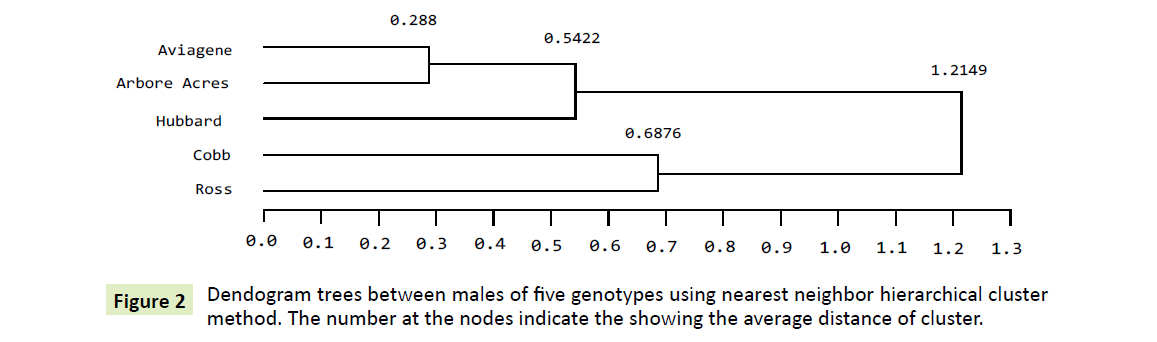
Phylogenetic tree for the five males: The dendogram clustering procedures splits the genetic divergence between the five genotypes males in a three distinct clusters. The first cluster grouped Aviagene and Arbor Acre males as the nearest homologous group confirming having a significant similar pattern for both as revealed in Table 2. Also they both recorded a smallest non-significant ΔCT least squares means estimate (-0.293). Their inter cluster distance was lowest among all clusters (0.288).
Hubbard males, the second one, had merged to the nearest one Aviagene and Arbor Acres recording node distance at (0.5422). Cobb and Ross males groups are merged in the third cluster although their ΔCT least squares means are not significantly the same. The intra cluster distance was found to be (0.6876) reflects a high variation exists and low gene flow among them [30]. Finally, maximum divergence between five male’s genotypes was shown by the dendogram clustering at degree of distance (1.2149).
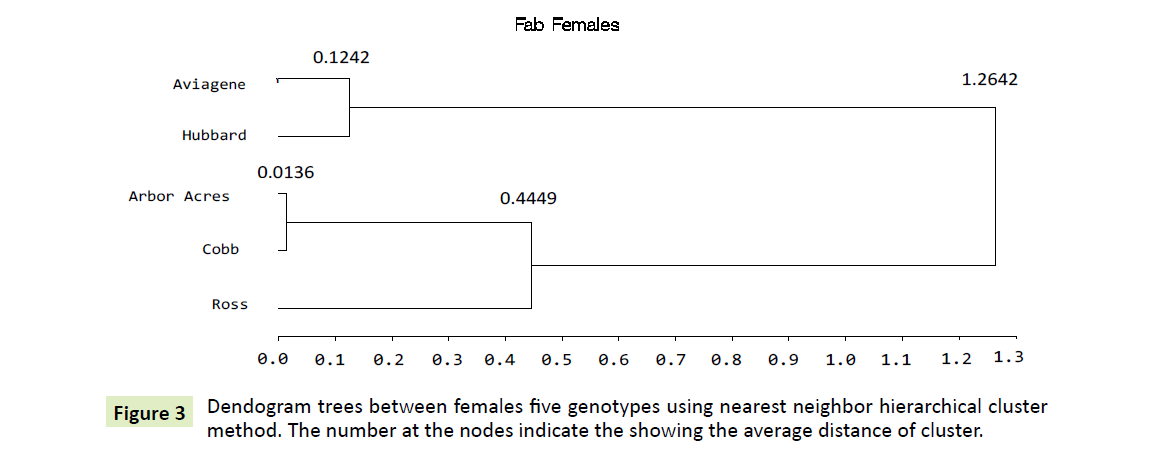
Phylogenetic tree for the five females: As shown in Figure 3, three distinct subgroups had been aggregated the five population groups. The Arbor Acre females had joined to Cobb at minimum divergence distance of (0.0136). Aviagen and Hubbard had compressed in the second one in an inter class distance of (0.1242). Ross, the third one, had merged to the nearest one, Arbor Acres and Cobb females at (0.4449) degree of distance. Finally, maximum divergence between five female’s genotypes was shown by the dendogram clustering at degree of distance (1.2642).
Significance of sex difference
Analysis of variance and contrasts: Aviagene, the unique genotype that their females possess significantly lower ΔCT least squares means over than males (14.00 vs. 14.56) (higher expression over males). Other genotypes showed significantly higher ΔCT least squares means for females over males. Hubbard genotype showed the lowest contrast estimate coefficient of females than males of all sex comparisons (0.153). It is the only genotype that their females observe non-significant (p=0.4079) superiority over males as shown in Table 4.
Table 4: Females contrast versus males for AFBP mRNA ∆CT (linear function ± SE) for five genotypes.
| Females vs. the rest | p-Value | F-Value | Contrast SS | Estimate | ∆CT | Genotype | |||
|---|---|---|---|---|---|---|---|---|---|
| p-Value | F-Value | Contrast SS | Estimate | ||||||
| 0.0493 | 4.22 | 2.3611518 | -0.280 ± 0.1365 | 0.0493 | 4.22 | 2.36115184 | -0.561 ± 0.273 | 14.00 ± 0.193 | Aviagen |
| 14.56 ± 0.193 | |||||||||
| 0.0291 | 5.29 | 16.95921 | 0.751 ± 0.326 | 0.0291 | 5.29 | 16.9592101 | 1.503 ± 0.653 | 15.77 ± 0.462 | Arbor Acre |
| 14.27 ± 0.462 | |||||||||
| 0.4079 | 0.71 | 0.70711 | 0.153 ± 0.182 | 0.4079 | 0.71 | 0.70710996 | 0.307 ± 0.365 | 14.19 ± 0.258 | Hubbard |
| 13.89 ± 0.258 | |||||||||
| 0.0019 | 11.86 | 4.6805514 | 0.401 ± 0.116 | 0.0019 | 11.86 | 4.68055142 | 0.803 ± 0.233 | 15.85 ± 0.167 | Cobb |
| 15.04 ± 0.162 | |||||||||
| 0.0286 | 5.35 | 4.6457138 | 0.400 ± 0.173 | 0.0286 | 5.35 | 4.64571383 | 0.800 ± 0.346 | 16.55 ± 0.249 | Ross |
| 16.74 ± 0.241 | |||||||||
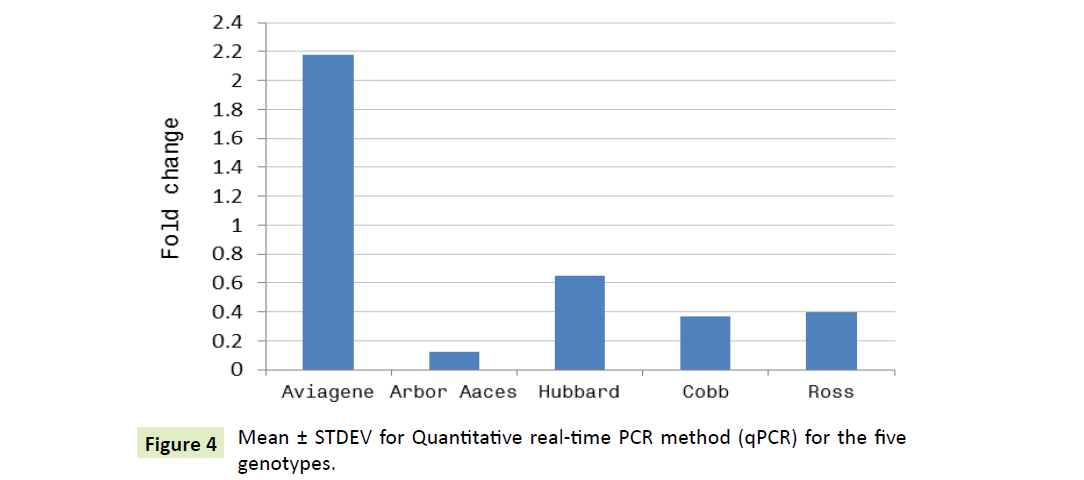
Fold change profile of fab gene for females over males
For each genotype, the gene expression profile of male genotype was used as the calibrator and fold change analyses is shown in Figure 4. Depending on Table 5, the expression of the A-FABP, only females of Aviagene genotype that express 2.176 fold more than males. Otherwise, Hubbard population, females possess 0.653 fold as males. Females of Ross, Cobb possess 0.395, 0.374, respectively. Arbor Acres females had the lowest 0.123 fold as their males. Consequently, both Aviagene and Hubbard females are nested in one cluster as shown in Figure 3.
Table 5: Gherlin mRNA ∆CT ± STDEV for five genotypes and their Fold change expression calculated by ΔΔCT method.
| Genotype | ∆∆CT ∆CT Females-∆CT Males |
Average ∆∆CT | Fold Change |
|---|---|---|---|
| Aviagen | -0.5612 ± 3.942 | -1.1222 (-4.503 to 3.381) |
2.176751765 |
| Arbor Acres | 1.5037 ± 3.655 | 3.007 (-2.152 to 5.159) |
0.124353927 |
| Hubbard | 0.3071 ± 4.126 | 0.614 (-3.819 to 4.433) |
0.653334942 |
| Cobb | 0.7125 ± 4.569 | 1.425 (-3.857 to 5.282) |
0.372431722 |
| Ross | 0.6697 ± 3.819 | 1.339 (-3.149 to 4.489) |
0.395187062 |
A-FABP and association with genotype, growth performance and carcass parameters
The A-FABP mRNA levels were significantly associated by genotype is consistent with that obtained by Li et al., (2008). Regarding to other FABP4 ΔCT genotypes means, higher FABP4 ΔCT mean (low expression) for Arbor Acres genotype appears to depress the development of growth performance and general carcass traits in Arbor Acre hybrid as given in Table 6. FABP4 ΔCT displayed significant negative association with live body weight of 37-day old (-83.03), fore muscle half weight (-35.35) and dorsal muscle half weight (-35.32), breast width (-0.33) carcass weight (-91.53), thigh muscle weight (-7.12), drum muscle weight (-3.72), major pectoralis muscles weight (-1.97) and small pectoralis muscles weight (-8.29).
Table 6: Association between A-FABP gene expression and phenotypic traits in both sexes of the commercial meat type chicken at 37-days old.
Values represent regression coefficient ±S.E. Values within a column significantly (P<0.05); LBW 37-d:Live Body Weight at 37- days; FH%:Fore Half%; DH:Dorsal Half; BW:Breast Width; CW:Carcass Weight; DMW:Drum Muscle Weight; TMW:Thigh Muscle Weight; MPMW:Major Pectoralis Muscle Weight; SPMW: Small Pectoralis Muscle Weight; LW: Liver Weight; HW: Heart Weight; GW: Gizzard Weight; HEW: Head Weight; HE&N: Head &neck; AF: Abdominal Fat; BRF: Breast Fat; THF: Thigh Fat; DRF:Drum Fat.
| Trait | Aviagen | P-Value | Arbor Acre | P-Value | Hubbard | P-Value | Cobb | P-Value | Ross | P-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| LBW 37-d | 24.16 ± 71.88 | 0.2869 | -83.03 ± 32.07 | 0.1071 | -32.64 ± 58.03 | 0.5913 | 112.84 ± 148.02 | 0.5637 | -61.05 ± 68.96 | 0.4054 |
| FH | -41.52 ± 37.46 | 0.3102 | -35.35 ± 12.35 | 0.0210 | -2.49 ± 18.27 | 0.8954 | 66.94 ± 55.73 | 0.2835 | -5.12 ± 33.48 | 0.8827 |
| FH% | -0.04 ± 0.014 | 0.0241 | 0.001 ± 0.007 | 0.255 | 0.008 ± 0.004 | 0.0571 | 0.04 ± 0.014 | 0.047 | 0.00 ± 0.00 | 0.4587 |
| DH | 8.87 ± 23.93 | 0.7237 | -35.32 ± 11.65 | 0.0163 | -7.98 ± 19.04 | 0.6878 | 13.75 ± 44.92 | 0.7719 | -24.00 ± 23.33 | 0.3377 |
| Bw | -0.04 ± 0.31 | 0.9112 | -0.33 ± 0.11 | 0.0151 | 0.19 ± 0.18 | 0.3269 | 0.58 ± 0.28 | 0.0916 | 0.056 ± 0.34 | 0.8718 |
| Cw | 11.45 ± 52.61 | 0.8349 | -91.53 ± 33.27 | 0.0250 | -26.065 ± 39.94 | 0.5348 | 55.06 ± 101.92 | 0.6122 | -21.23 ± 56.71 | 0.7192 |
| DMW | 3.86 ± 2.49 | 0.1727 | -3.72 ± 1.48 | 0.0363 | -0.45 ± 2.36 | 0.8536 | 2.56 ± 6.15 | 0.6943 | -1.58 ± 2.97 | 0.3714 |
| TMW | -1.41 ± 7.36 | 0.8541 | -7.12 ± 2.31 | 0.0151 | 0.07 ± 4.75 | 0.9875 | 3.14 ± 14.09 | 0.8326 | -4.15 ± 4.34 | 0.6112 |
| MPMW | 0.43 ± 1.74 | 0.812 | -1.97 ± 0.58 | 0.0094 | -1.04 ± 1.04 | 0.4151 | 2.61 ± 2.78 | 0.3893 | -0.22 ± 1.21 | 0.9822 |
| SPMW | -1.83 ± 6.92 | 0.8001 | -8.29 ± 3.33 | 0.0375 | -3.39 ± 3.86 | 0.1879 | 9.14 ± 12.98 | 0.5127 | -0.24 ± 1.53 | 0.8635 |
| Lw% | 0.004 ± 0.003 | 0.2016 | 0.00 ± 0.001 | 0.576 | -0.001 ± 0.003 | 0.7153 | -.008 ± 0.003 | 0.0275 | -0.001 ± 0.002 | 0.5401 |
| SpW% | -0.00 ± 0.00 | 0.6921 | 0.0003 ± 0.0001 | 0.0221 | 0.000 ± 0.000 | 0.5135 | 0.00 ± 0.000 | 0.8812 | -0.000 ± 0.000 | 0.2488 |
| LW | -1.83 ± 4.79 | 0.7161 | -1.52 ± 0.98 | 0.1607 | -0.84 ± 0.75 | 0.0447 | 8.08 ± 3.91 | 0.0936 | -1.27 ± 3.53 | 0.7295 |
| LW% | -0.001 ± 0.003 | 0.6738 | 0.002 ± 0.001 | 0.0381 | -0.000 ± 0.000 | 0.4057 | 0.006 ± 0.004 | 0.2485 | -0.000 ± 0.002 | 0.8240 |
| HW | 1.17 ± 0.41 | 0.0305 | -0.32 ± 0.21 | 0.1569 | -.077 ± 0.41 | 0.1009 | -0.14 ± 0.88 | 0.8830 | -0.098 ± 0.54 | 0.8593 |
| HW% | 0.0009 ± 0.0003 | 0.0139 | 0.00 ± 0.00 | 0.2635 | -0.000 ± 0.000 | 0.1221 | -0.000 ±0.000 | 0.5258 | 0.000 ± .0002 | 0.824 |
| GW% | 0.00 ± 0.00 | 0.4434 | 0.00 ± 0.00 | 0.0536 | 0.000 ± 0.000 | 0.7114 | -0.002 ± 0.000 | 0.0431 | -0.000 ± 0.001 | 0.9463 |
| HEW | 3.76 ± 2.61 | 0.1995 | 0.002 ± 0.001 | 0.4127 | 0.85 ± 1.69 | 0.6285 | -1.95 ± 4.02 | 0.6476 | -1.74 ± 3.79 | 0.6592 |
| HE&N% | 0.003 ± 0.002 | 0.3744 | 0.004 ± 0.001 | 0.003 | 0.000 ± 0.002 | 0.9141 | 0.001 ± 0.009 | 0.8807 | -0.001 ± 0.003 | 0.6227 |
| AF | -3.84 ± 4.53 | 0.4294 | -2.51 ± 1.19 | 0.0681 | -0.93 ±3.42 | 0.7938 | 17.07 ± 4.34 | 0.011 | -1.01 ± 2.77 | 0.7265 |
| AF% | -0.004 ± 0.004 | 0.3428 | 0.00 ± 0.001 | 0.86 | -0.000 ± 0.003 | 0.9326 | 0.014 ± 0.003 | 0.0062 | -000 ± 0.002 | 0.6974 |
| BRF% | -0.03 ± 0.04 | 0.4769 | 0.013 ± 0.04 | 0.7149 | 0.05 ± 0.60 | 0.4475 | -0.19 ± 0.10 | 0.1056 | 0.02 ± 0.06 | 0.6784 |
| THF% | -0.18 ± 0.13 | 0.2368 | 0.07 ± 0.08 | 0.3853 | -0.04 ± 0.33 | 0.9108 | -1.72 ± 0.80 | 0.0840 | -0.12 ± 0.24 | 0.6234 |
| DRF% | 2.95 ± 1.38 | 0.0764 | -0.11 ± 0.40 | 0.7980 | -0.08 ± 0.90 | 0.2661 | -0.30 ± 0.64 | 0.7743 | 0.086 ± 0.14 | 0.5651 |
At young slaughter age (37days old) and growing ration, Cobb males seemed to be sensitive at low gene expression as presented in Table 7. It was denoted that many growth performance and carcass traits were significantly (p<0.05) had positive association higher FABP4 ΔCT mean (low expression) such as live body weight of 21 days old (166.41), live body weight of 28 days old (397.40), live body weight of 35 days (605.61), weight gain of 0-21 days (162.81), specific growth rate of 0-21 days (0.01), specific growth rate of 0-37 days (0.01), growth efficiency of 0-21 days (3.07), and growth efficiency 0-37 days (15.20). High abdominal fat accumulation may responsible for elevation for live body weight parameters in male Cobb hybrids. The study of Chen et al. observed that the A-FABP transcript levels are increased rapidly with the body weight in pigs until 60-70 kg and lasted at high levels in both breeds studied and this may be due to the elevated IMF (intramuscular fat content) and high marbling in pigs, which are responsible for increasing body weight, and not due to the muscular growth itself [18]. Emphasizing this hypothesis, Nafikov et al. reported that certain FABP4 haplotypes have an association with particular fatty acid profiles in milk without differences in milk yield in cattle [23].
Table 7: Association between A-FABP gene expression and phenotypic traits in the commercial meat type male chicken at 37-days old.
Values represent regression coefficient ±S.E. Values within a column significantly (P<0.05); LBW 21, 28, 35-d:Live Body Weight at 21, 28, 35-days; WG 0-21:Weight Gain 0-21 day old; SGR 0-21, 0-37:Specific Growth Rate 0-21, 0-37 days old; SpW:Spleen Weight; HW: Heart Weight; AFW: Abdominal Fat Weight; BMF: Breast Muscle Fat; TMF: Thigh Muscle Fat; DMF%:Drum Muscle Fat%.
| Trait | Aviagen | P-Value | Arbor Acre | p-value | Hubbard | p-value | Cobb | p-value | Ross | P-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| LBW 21-d | -29.74 ± 52.45 | 0.6279 | 36.70 ± 33.06 | 0.3479 | -21.90±39.97 | 0.6218 | 166.41 ± 36.94 | 0.0459 | 40.87 ± 55.79 | 0.5169 |
| LBW 28-d | -25.04 ± 93.35 | 0.8137 | 38.63 ± 60.71 | 0.5698 | -51.19±59.88 | 0.4555 | 397.40 ± 8.79 | 0.0465 | 81.01 ± 67.56 | 0.3676 |
| LBW 35-d | -149.14 ± 123.36 | 0.3502 | 34.93 ± 30.97 | 0.3414 | -57.44±73.75 | 0.4929 | 605.61 ± 23.59 | 0.0392 | 96.23 ± 91.42 | 0.3698 |
| WG 0-21 | 27.99 ± 51.55 | 0.6416 | 34.84 ± 32.91 | 0.3674 | -17.49±39.88 | 0.6907 | 162.81 ± 33.72 | 0.0403 | 39.66 ±55.60 | 0.5272 |
| SGR 0-21 | 0.00 ± 0.004 | 0.9739 | 0.001 ± 0.002 | 0.6947 | 0.003±0.004 | 0.5274 | 0.01 ± 0.002 | 0.0426 | 0.002 ± 0.005 | 0.7123 |
| SGR 0-37 | -0.005 ± 0.001 | 0.0096 | -0.00 ± 0.00 | 0.7309 | 0.00±0.001 | 0.5048 | 0.01 ± 0.002 | 0.0429 | 0.00 ± 0.003 | 0.9875 |
| GE 021 | 0.05 ± 1.28 | 0.9737 | 0.29 ± 0.65 | 0.6876 | 0.69± 0.91 | 0.5048 | 3.07 ± 0.66 | 0.0435 | 0.52 ± 1.19 | 0.691 |
| GE037 | -6.48 ± 0.87 | 0.0176 | -0.45 ± 1.23 | 0.7407 | 2.66±1.99 | 0.2725 | 15.20 ± 3.39 | 0.0463 | 0.06 ± 3.54 | 0.9867 |
| SpW | -1.22 ± 0.35 | 0.0717 | 0.12 ± 0.33 | 0.7362 | 0.27 ± 0.29 | 0.419 | 1.89 ± 0.18 | 0.0087 | -0.31 ± 0.20 | 0.2232 |
| HW | 0.59 ± 0.41 | 0.2904 | 0.40 ± 0.27 | 0.2329 | -0.93± 0.24 | 0.0298 | 3.09 ± 1.21 | 0.1263 | -0.18 ± 1.12 | 0.882 |
| AFW | -8.66 ± 7.37 | 0.3608 | 1.28 ± 2.92 | 0.6905 | -3.65 ±2.43 | 0.2310 | 34.03 ± 4.05 | 0.0139 | 7.07 ± 5.30 | 0.9867 |
| BMF% | -0.14 ± 0.03 | 0.0522 | -0.04 ± 0.05 | 0.4992 | 0.04±0.11 | 0.7319 | -0.11 ± 0.30 | 0.7444 | -0.12 ± 0.10 | 0.3189 |
| TMF% | -0.07 ± 0.21 | 0.7782 | 0.27 ± 0.32 | 0.4555 | 0.37±0.58 | 0.5728 | -3.82 ± 1.20 | 0.0857 | -0.35 ± 0.37 | 0.4146 |
| DMF% | 5.10 ± 2.71 | 0.2002 | 1.33 ± 1.23 | 0.3588 | -0.74±0.90 | 0.0549 | 1.59 ± 1.65 | 0.4361 | 0.10 ± 0.33 | 0.7894 |
Ross females showed significant positive association of FABP4 ΔCT on specific growth rate and growth efficiency at 21-37 days as in Table 8. Other fluctuated effects were shown among other genotypes. FABP4 ΔCT mean for Hubbard genotype observed significant (p<0.05) negative association with liver weight (-0.84) (Table 6) and negative association with heart weight (-0.93) for males (Table 7). Aviagene genotype, both sexes (Table 6), a significant negative correlation (-0.04) of FABP4 ΔCT mean with half percentage of the fore muscle weight and positive association with the heart weight (1.17) was found. Difficult ability to detect significant associations between FABP4 and quantitative nature of growth parameters for commercial broilers is not only controlled by many genes and environmental factors but also broad variety of populations of different origins and breeding history [31,32].
Table 8: Association between A-FABP gene expression and phenotypic traits in the commercial meat type female chicken at 37-days old.
Values represent regression coefficient ±S.E. Values within a Column Significantly (P<0.05); SGR 21-37:Specific Growth Rate 21-37days; GE 21-37:Growth Efficiency 21-37 days; MPMW: Major Pectoralis Muscle Weight; LL: Leg Length; LW: leg Weight; WW: Wing Weight; HW: Heart Weight; HE&N: Head&neck; AFW: Abdominal Fat Weight; BMF: Breast Muscle Fat; TMF: Thigh Muscle Fat; DMF%:Drum Muscle Fat%.
| Trait | Aviagen | P-Value | Arbor acre | P-Value | Hubbard | P-Value | Cobb | P-Value | Ross | P-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| SGR21-37 | -0.005 ± 0.007 | 0.5347 | -0.001 ± 0.001 | 0.3769 | -0.006 ± 0.005 | 0.3494 | 0.01 ± 0.01 | 0.4645 | 0.003 ± 0.0003 | 0.0148 |
| GE 21-37 | -0.17 ± 0.30 | 0.5711 | -0.05 ± 0.04 | 0.3680 | -0.24 ± 0.19 | 0.3283 | 0.31 ± 0.29 | 0.4795 | 0.10 ± 0.01 | 0.0118 |
| MPMW% | -0.003 ± 0.003 | 0.4260 | -0.001 ± 0.0002 | 0.0130 | 0.001 ± 0.006 | 0.9229 | -0.005 ± 0.01 | 0.4815 | 0.006 ± 0.009 | 0.5241 |
| L L | -0.03 ± 0.21 | 0.9093 | -0.052 ± 0.010 | 0.6520 | 0.21 ± 0.08 | 0.1107 | -0.75 ± 0.20 | 0.1644 | 0.11 ± 0.003 | 0.001 |
| L W % | 0.001 ±0.001 | 0.603 | 0.001 ± 0.001 | 0.1653 | -0.001 ± 0.002 | 0.5583 | -0.01 ± 0.001 | 0.0489 | -0.001 ± 0.001 | 0.4335 |
| WW % | -0.01 ± 0.01 | 0.2288 | 0.004 ± 0.002 | 0.1443 | 0.003 ± 0.001 | 0.0217 | -0.01 ± 0.001 | 0.1851 | 0.00 ± 0.004 | 0.9368 |
| SpW | 0.54 ± 0.11 | 0.0379 | 0.20 ± 0.15 | 0.2745 | -0.012 ± 0.23 | 0.9502 | -0.52 ± 0.09 | 0.1065 | 0.002 ± 0.21 | 0.9924 |
| H W | 0.85 ± 0.09 | 0.0108 | 0.0001 ± 0.0001 | 0.3723 | -0.001 ± 0.001 | 0.5744 | -0.00 ± 0.00 | 0.8935 | 0.0003 ± 0.0001 | 0.2823 |
| H W% | 0.00 ± 0.001 | 0.072 | 0.002 ± 0.001 | 0.3048 | 0.001 ± 0.004 | 0.4903 | -0.001 ± 0.003 | 0.9194 | -0.001 ± 0.002 | 0.2121 |
| HE & N% | 0.005 ± 0.01 | 0.5321 | 0.004 ± 0.001 | 0.011 | 0.001 ± 0.002 | 0.6701 | 0.002 ± 0.001 | 0.5925 | -0.003 ± 0.004 | 0.5732 |
| AFW | 6.48 ± 3.63 | 0.2165 | -3.51 ± 1.59 | 0.1145 | -1.01 ± 3.47 | 0.7977 | 14.88 ± 4.42 | 0.1837 | -3.40 ± 0.60 | 0.0294 |
| B M F% | 0.04 ± 0.05 | 0.5009 | 0.04 ± 0.05 | 0.4909 | 0.04 ± 0.09 | 0.6657 | -0.10 ± 0.05 | 0.2709 | 0.06 ± 0.05 | 0.3451 |
| T M F% | -0.07 ± 0.17 | 0.7292 | 0.08 ± 0.04 | 0.1145 | -0.19 ± 0.31 | 0.5980 | 0.31 ± 0.30 | 0.4904 | -0.07 ± 0.46 | 0.8964 |
| D M F% | 0.38 ± 0.95 | 0.7276 | 0.002 ± 0.26 | 0.9939 | -0.48 ± 0.30 | 0.2515 | -0.30 ± 0.10 | 0.2034 | -0.01 ± 0.12 | 0.9458 |
Association of A-FABP gene expression with fat accumulation
Fat deposition in chickens was basically occurred in visceral adipose tissue and muscles, particularly the intramuscular fat content (IMF). The results demonstrated that FABP4 ΔCT mean displayed positive significant association with abdominal fat (17.07) and abdominal fat percentage (0.014 g) only in Cobb genotype (Table 6) and significant positive correlation with abdominal fat in their males (34 g) (Table 7). As previously noticed at Table 4 and Figure 1, they had significantly higher mRNA expression than females. Negative association of FABP4 ΔCT mean with abdominal fat weight (-3.40). Table 8 was notice for Ross females although they had FABP4 mRNA expression lower than males (Table 4 and Figure 1). As reported by Li et al. [33], The A-FABP gene expression is affected by gender. Only Aviagen females that had significantly higher A-FABP gene expression than males (Table 4 and Figure 1). Results herein showed that there was no association with fat accumulation in abdomen or in muscle for all other hybrids. Nevertheless, the non-significance effect of the A-FABP gene transcription on the intramuscular fat content percentage (IMF) in breast, thigh and drum muscles of all of the five genotypes is consistent with the findings of Ye et al. [20] who found that the A-FABP gene mRNA expression level is positively correlated significantly with abdominal fat but not with IMF content in Rugao and Luyuan chickens. Finally, the non-specificity of the A-FABP gene for abdomen fat and IMF content for these hybrids has many reasons. Firstly, it may be due to the same moderate calorie content of ration for these hybrids (growing ration), where feeding strategy is necessary to alter intramuscular fat profile in meat through manipulating gene expression of enzymes related to fat accumulation [34]. The finding of Saez et al. [35] who found that the A-FABP protein content in pectoralis major (PM) muscle of ducks was not significantly affected by dietary level for each nutritional condition, have to be emphasized. Secondly, the young slaughter age (37 day) as a broiler where age is a strong factor for fat accumulation. Third, broilers are selected for high growth traits not fatness. In marbled pork production, it is known that production is characterized by largely elevated IMF with higher A-FABP transcript levels in muscle of fatty pig breed compared to the leaner ones where Fatty acids are transported in fewer quantities through intracellular trafficking in leaner breeds resulting in less IMF deposits than fatty ones [18,21,36]. Therefore, an association between DNA polymorphisms in the A-FABP gene and fat accumulation in chickens is reported [37] and pigs [38,39]. In exon 1, a substitution mutation is significantly associated with abdominal fat, subcutaneous fat and intramuscular fat content of chicken [40]. Also, a new G/A polymorphism in exon 3 of the chicken A-FABP gene is associated with abdominal fat percentage [41,42].
Conclusion
Genetic divergence for A-FABP gene quantity among five commercial hybrids was achieved using clustering analysis hierarchical method. Cobb and Ross genotypes were much closer to each other and the same for both Arbor Acres and Hubbard at further distance. Aviagen genotype is unique but much closer to Arbor Acres and Hubbard genotype. Little abundance of mRNA A-FABP gene is responsible for growth performance depression in Arbor Acre genotype and responsible for strongly positive association with growth performance for the Cobb males. A-FABP gene has no effect on total muscle fat% content for all genotypes and can manifest a potential use in advanced molecular research to heal the excess of abdominal fat in Cobb genotype.
References
- Griffin HD (1996) Understanding genetic variation in fatness in chickens. Annual report Roslin Institute Edinburgh.
- Jennen DGJ, Vereijken ALJ, Bovenhuis H, Crooijmans R, Veenendaal A, et al. (2004) Detection and localization of quantitative trait loci affecting fatness in broilers. Poult Sci 83: 295-301.
- Yuan JM, Guo YM, Yang Y, Wang Z (2007) Characterization of fatty acid digestion of Beijing fatty and arbor acres chickens. Asian-Aust J Anim Sci 20: 1222-1228.
- Rothschild MF, Soller M (1997) Candidate gene analysis to detect genes controlling traits of economic importance in domestic livestock. Probe 8: 13-20.
- Li CC, Li K, Li J, Mo DL, Xu RF, et al. (2006) Polymorphism of ghrelin gene in twelve Chinese indigenous chicken breeds and its relationship with chicken growth traits. Asian-Aust J Anim Sci 19: 153-159.
- Oh JD, Kong HS, Lee JH, Choi IS, Lee SJ, et al. (2006) Identification of novel SNPs with effect on economic traits in uncoupling protein gene of Korean native chicken. Asian-Aust J Anim Sci 19: 1065-1070.
- Wang Q, Li H, Li N, Leng L, Wang Y, et al. (2006) Identification of single nucleotide polymorphism of adipocyte fatty acid-binding protein gene and its association with fatness traits in the chicken. Poult Sci 85: 429-434.
- Chmurzyńska A (2006) The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Genet 47: 39-48.
- Michal JJ, Zhang ZW, Gaskins CT, Jiang Z (2006) The bovine fatty acid binding protein 4 gene is significantly associated with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Anim Genet 37: 400-402.
- Weisiger RA (2002) Cytosolic fatty acid binding proteins catalyze two distinct steps in intracellular transport of their ligands. Mol Cell Biochem 239: 35-43.
- Zimmerman AW, Veerkamp JH (2002) New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci 59: 1096-1116.
- Hertzel AV, Bernlohr DA (2000) The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol Metab 11: 175-180.
- Brandstetter AM, Sauerwein H, Veerkamp JH, Geay, Y, Hocquette JF (2002) Effects of muscle type, castration, age and growth rate on H-FABP expression in bovine skeletal muscle. Livest Sci 75: 199-208.
- Furuhashi M, Hotamisligil GS (2008) Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 7: 489-503.
- Gerbens F, Jansen A, van Erp AJM, Harders, F, Meuwissen, et al. (1998) The adipocyte fatty acid-binding protein locus: characterization and association with intramuscular fat content in pigs. Mamm Genome 9: 1022-1026.
- Hertzel AV, Smith LA, Berg AH, Cline GW, Shulman GI, et al. (2006) Lipid metabolism and adipokine levels in fatty acid-binding protein null and transgenic mice. Am J Physiol Endocrinol Metab. 290: E814-E823.
- Xu Qn L, Tang GW, Zhang QL, Huang YK, Liu YX, et al. (2011) The FABP4 gene polymorphism is associated with meat tenderness in three Chinese native sheep breeds. Czech J Anim Sci 56: 1-6.
- Chen QM, Wang H, Zeng YQ, ChenW (2013) Developmental changes and effect on intramuscular fat content of H-FABP and A-FABP mRNA expression in pigs J Appl Genet 54: 119-123.
- Aviles C, Polvillo O, Pena F, Juarez M, Martinez AL, et al. (2013) Associations between DGAT1, FABP4, LEP, RORC, and SCD1 gene polymorphisms and fat deposition in Spanish commercial beef. J Anim Sci 91: 4571-4577.
- Ye MH, Chen JL, Zhao GP, Zheng MQ, Wen J (2010) Associations of A-FABP and H-FABP markers with the content of intramuscular fat in Beijing-You chicken. Anim Biotechnol 21: 14-24.
- Gao Y, Zhang YH, Zhang S, Li F, Wang S, et al. (2011) Association of A-FABP gene polymorphism in intron 1 with meat quality traits in Junmu No. 1 white swine. Gene 487: 170-173.
- Shin SC, Heo JP, Chung ER (2012) Genetic variants of the FABP4 gene are associated with marbling scores and meat quality grades in Hanwoo (Korean cattle). Mol Biol Rep 39: 5323-5330.
- Nafikov RA, Schoonmaker JP, Korn KT, Noack K, Garrick DJ, et al. (2013) Association of polymorphisms in solute carrier family 27, isoform A6 (SLC27A6) and fatty acid-binding protein-3 and fatty acid-binding protein-4 (FABP3 and FABP4) with fatty acid composition of bovine milk. J Dairy Sci 96: 6007-6021.
- Liu ZW, Fan HL, Liu XF, Ding XB, Wang T, et al. (2015) Overexpression of the A-FABP gene facilitates intermuscular fat deposition in transgenic mice. Genet Mol Res 14: 2742-2749.
- Shu L, Hoo RLC, Wu X, Pan Y, Lee IPC, et al. (2017) A-FABP mediates adaptive thermogenesis by promoting intracellular activation of thyroid hormones in brown adipocytes. Nat Commun 8: 1-16.
- Gondwe TNP, Wollny CBA (2005) Evaluation of the growth potential of local chickens in Malawi. International Journal of Poultry Science 4: 64-70.
- Davis MJ (2010) Contrast Coding in Multiple Regression Analysis: Strengths, Weaknesses, and Utility of Popular Coding Structures. J Data Sci 8: 61-73.
- Abdi H, Williams L (2010) Contrast Analysis: In Encyclopedia of Research Design, edited by Salkind NJ, Dougherty DM, and Frey B. Thousand Oaks: Sage 243-51.
- Shavelson RJ (2016) Statistical Reasoning for the Behavioral Sciences translated by Güler N Ankara: PegemA.
- Yakubu A and Ugbo SB (2011) An assessment of biodiversity in morphological traits of Muscovy ducks in Nigeria using discriminant analysis. Proceedings of 2010 International Conference on Biology Environment and Chemistry, IACSIT Press, Singapore pp. 389-391.
- Andersson L (2001) Genetic dissection of phenotypic diversity in farm animals. Nat Rev Genet 2: 130-138.
- Blecha IMZ, Siqueira F, Ferreira ABR, Feijó GLD, Junior RAAT, et al. (2015) Identification and evaluation of polymorphisms in FABP3 and FABP4 in beef cattle. Genet Mol Res 14: 16353-16363.
- Li WJ, Li HB, Chen JL, Zhao GP, Zheng MQ, et al. (2008) Gene Expression of Heart- and Adipocyte-Fatty Acid-Binding Protein and Correlation With Intramuscular Fat in Chinese Chickens. Anim Biotechnol 19: 189-193.
- Dervishi E, Serrano C, Joy M, Serrano M, Rodellar C, et al. (2011). The effect of feeding system in the expression of genes related with fat metabolism in semitendinous muscle in sheep. Meat Sci. 89: 91-97.
- Saez G, Davail S, Gentes G, Hocquette J F, Jourdan T, et al. (2009) Gene expression and protein content in relation to intramuscular fat content in Muscovy and Pekin ducks. Poult Sci 88: 2382-2391.
- Jacyno E, Pietruszka A, Kawecka M, Biel W, Kotodziej-Skalska A (2015) Phenotypic correlations of backfat thickness with meatiness traits, intramuscular fat, longissimus muscle cholesterol and fatty acid composition in pigs. S Afr J Anim Sci 45: 122-128.
- Chen KW, Zhang SJ, Tang QP, Gao YS, Li HF (2006) Analysis on Genetic Variation of Adipocyte Fatty Acid Binding Protein Gene in Different Chicken Breeds. acta veterinaria et zootechnica sinica, 37: 1114-1114.
- Gerbens F, van Erp AJ, Harders FL, Verburg FJ, Meuwissen TH, et al. (1999). Effect of genetic variants of the heart fatty acid-binding protein gene on intramuscular fat and performance traits in pigs. J Anim Sci, 77: 846-852.
- Gerbens F, Verburg F J, van Moerkerk H T, Engel B, Buist W, et al. (2001). Association of heart and adipocyte fatty acid-binding protein gene expression with intramuscular fat content in pigs. J Anim Sci 79: 347-354.
- Luo GF, Chen JL, Wen J, Zhao GP, Zheng MQ, et al. (2006) Study of single nucleotide polymorphism of A-FABP gene and its association with fatness traits in chicken. Yi chun 28: 39-42.
- Wang Q, Guan T, Li H, Bernlohr DA (2009) A Novel Polymorphism In The Chicken Adipocyte Fatty Acid-Binding Protein Gene (FABP4) That Alters Ligand-Binding and Correlates With Fatness. Comp Biochem Physiol B Biochem Mol Biol 154: 298-302.
- Ye MH, Chen JL, Zhao GP, Zheng MQ, Wen J (2008) Associations of A-FABP and H-FABP markers with the content of intramuscular fat in Beijing-you chickens. Front Agric China 2: 474–479.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences