ISSN : 2249 - 7412
Asian Journal of Plant Science & Research
Estimates of Genetic Parameters and Agronomic Variability of Nutsedge (Cyperus esculentus L.) Ecotypes Collected in Niger
Bori Haoua1, Idi Saidou Sani2,5*, Ali Bibata1, Dan Guimbo Iro3, Adam Toudou3 and Bakasso Yacoubou4,5
1Department of Irrigated Crops, National Institute of Agronomic Research of Niger, Niamey, Niger
2Department of Plant Production and Biodiversity, Faculty of Environment Science, University of Diffa, Diffa, Niger
3Faculty of Agronomy, Abdou Moumouni University, Niamey, Niger
4Department of Biology, Faculty of Science and Techniques, Abdou Moumouni University, Niamey, Niger
5Laboratory for the Management and Valorization of Biodiversity in the Sahel (GeVaBioS), Abdou Moumouni University, Niamey, Niger
- *Corresponding Author:
- Idi Saidou Sani
Department of Plant Production and Biodiversity, Faculty of Environment Science, University of Diffa, Diffa,
Niger,
E-mail: saidousani@yahoo.fr
Received date: March 23, 2024, Manuscript No. AJPSKY-24-18794; Editor assigned date: March 26, 2024, PreQC No. AJPSKY-24-18794 (PQ); Reviewed date: April 09, 2024, QC No. AJPSKY-24-18794; Revised date: April 16, 2024, Manuscript No. AJPSKY-24-18794 (R); Published date: April 23, 2024, DOI: 10.36648/2249-7412.14.2.313
Citation: Haoua B, Sani IS, Bibata A, Iro DG, Toudou A, et al. (2024) Estimates of Genetic Parameters and Agronomic Variability of Nutsedge (Cyperus esculentus L.) Ecotypes Collected in Niger. Asian J Plant Sci Vol:14 No:2: 313.
Abstract
The work for estimation of genetic characters would be useful in developing appropriate selection and strategies. Heritability is a measure of possible genetic improvement and advancement under selection. Aim of the study was to determine the level of agro-morphological diversity of Niger nutsedge ecotypes and to predict which parameters have good genetic heritability within the collection, with a view to identifying improvement strategies. The plant material consists of five ecotypes of Niger nutsedge, four of which are cultivated and one wild. The agromorphological characterization took place during the June 2017 rainy season on the experimental field of the Faculty of Agronomy. The results of the study carried out on the ecotypes using a five-repeat Fisher block design revealed significant morphological diversity structured around vegetative traits and yield and a division of the ecotypes into three groups based mainly on Plant Height (HP), Tuber Emulsion Length (TEL) and tuber yield (RdT). Evaluation of the genetic parameters showed that low phenotypic and genotypic coefficients of variation were observed for the characters number of floral branches (8.851% and 9.329%). The heritability of the traits studied, ranging from 35.544% to 99.358%, is high for all traits. The expected genetic gain relative to the trait mean (GA (% trait mean)) ranged from 16.697% for the width of the second leaf blade to 99.816% for the diameter of the mother plant. The three groups obtained could be used as breeding stock for the creation of varieties that meet farmers' expectations. If the selection objective is yield, it could be oriented towards group 1 and 2 ecotypes, which have the best tuber yields. But if the selection objective is nutritional, the choice could be directed towards group three, whose are very rich in iron. The genetic diversity observed within Niger nutsedge ecotypes could be exploited in breeding programs.
Keywords
Ecotype; Nutsedge; Agronomic; Genetic variability
Introduction
Yellow nutsedge (Cyperus esculentus L.) is a plant in the Cyperaceae family [1]. The nutsedge plant produces edible tubers at maturity, commonly known as "sweet peas" [2]. Nutsedge is highly productive, with yields ranging from over 5 t/ha to 14 Tha-1 in farmers' fields [3,4]. In Niger, the development of yellow nutsedge cultivation only really got underway in 1985 [5]. Nutsedge (Cyperus esculentus) is a plastic species and this plasticity can have consequences for the intra-specific determination of specimens [6]. The effect of the environment can profoundly alter the plant's appearance. Plasticity can also easily lead to a wide variety of morphological forms resulting from interactions between the environment and different genotypes [6].This makes precise variety identification difficult [7]. Molecular biology methods make it possible to identify these species regardless of environmental variations. Genetic markers provide information on the genotype of an individual and are not modified by the environment [8,9]. They can be used throughout an experiment and can be observed at any stage of plant development and on any organ (the plant's genetic information is contained in its entirety in all cells). They can give results after a single collection of vegetative material from the field [10]. The study carried out by [11] on the agro-morphological characterization of Niger nutsedge ecotypes enabled us to distinguish them in terms of their morphological variability and agronomic potential. Studies by [12] revealed relatively high genetic diversity among Niger nutsedge ecotypes. The aim of this study is to assess the agro-morphological diversity of Niger nutsedge ecotypes and to estimate the genetic parameters in order to identify the best strategies for improving and exploiting this genetic resource.
Materials and Methods
Plant material
The plant material consisted of five ecotypes of Niger nutsedge, four of which are cultivated and one wild. Tubers of the cultivated nutsedge were collected from cereal markets in Maradi and Dosso. Wild nutsedge tubers were collected on uncultivated land in the village of Rijia Samna in the Dosso region. Ecotypes were selected on the basis of three criteria:
• Ecotype color
• Soil type in production areas
• The importance of cultivation of the two types of nutsedge in these production zones
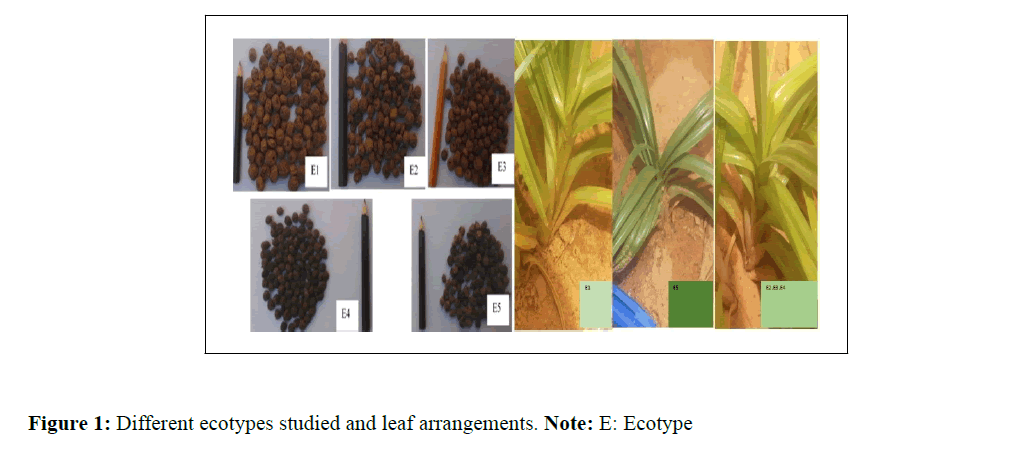
Then the different ecotypes studied are: Maradi nutsedge with large tubers, Maradi nutsedge with small black tubers and Dosso nutsedge with small tubers (Table 1) and (Figure 1).
| Characteristics | |||||
|---|---|---|---|---|---|
| Types | Ecotypes | NOMS | Colors | Forms | Province |
| GrS | E1 | Large nutsedge | Light brown | Round | Maradi |
| petS | E2 | Small nutsedge | Dark brown | Round | Maradi |
| petS | E3 | Red nutsedge | Red | Round | Dosso |
| petS | E4 | Small nutsedge | Black | Round | Maradi |
| petSS | E5 | Wild nutsedge | Black | Oval | Dosso |
| Note: GrS : Big nutsedge; petS: Small nutsedge; petSS: Small wild nutsedge; E: Ecotype. | |||||
Table 1: Characteristics of the Niger nutsedge ecotypes studied and their origin.
Experimental site
Agro-morphological characterization took place during June 2017 rainy season at the experimental site of the Faculty of Agronomy, Abdou Moumouni University, Niamey (13°29'58.7'' north latitude and 002°05'25.0'' east longitude) (Figure 2). The choice of this site was justified by the environmental conditions similar to those found in tiger nut-growing areas (sandy soils and rainfall of between 300 and 600 mm), to ensure better performance of the genotypes for the various agromorphological traits studied. Rainfall recorded over the last ten years varies from 500 to over 700 mm at the test site. The soil is characterized by a sandy to sandy-loam texture [11].
Methods
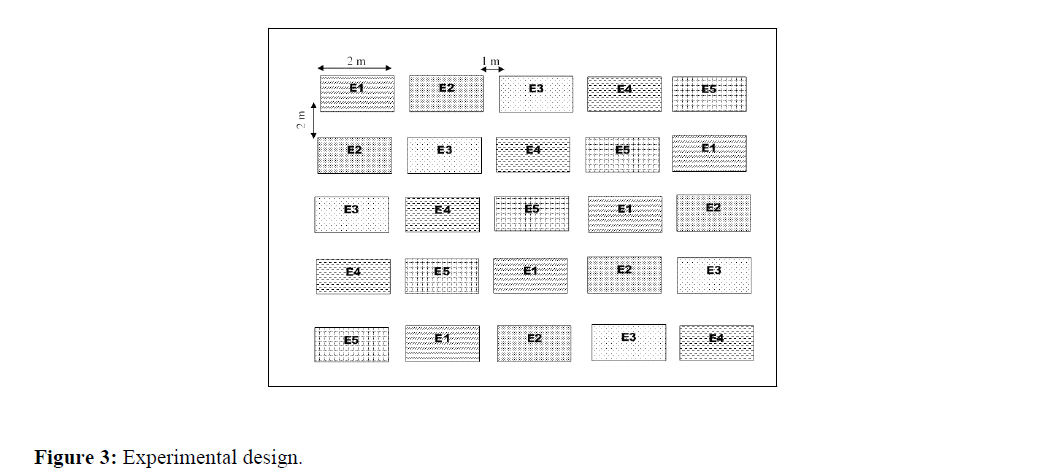
Experimental design: The trial was set-up over an area of 182 m2 using Fischer block design with 5 replicates (elementary plots) for each ecotype. The elementary plots have a surface area of 2 m2 (2 m × 1 m). Blocks are separated by a 2 m aisle, while two consecutive plots are 1 m apart. Within a plot, the spacing between crop rows is 20 cm. On the same cultivation line, bunches are spaced 20 cm apart. The crop density is 66 bunches per elementary plot (Figure 3).
Parameters measured: Sixteen quantitative characteristics were used to characterize the different ecotypes. The length of the first leaf blade (LPL), the length of the second leaf blade (LDL), the length of the third leaf blade (LTL), the width of the first leaf blade (LarPL), the width of the second leaf blade (LarDL), the width of the third leaf blade (LarTL), the Diameter of the Mother Plant (DPM), the Height of the Plant (HP), Number of Tubers Per bunch (NTP), Tuber Emission Length (TEL), Maximum Root Length (MRL), distance between mother plant and tillers (DPmT), number of flowering shoots (NHF), number of spikelets (NE), tuber yield (RdT) and biomass yield (RdB). With the exception of yields, which were determined using the 1 m × 1 m yield square method, the other characteristics were measured on 50 plants sampled per ecotype either10 plants per plot.
Analysis of collected data: GenStat v9.1, Minitab version 18 and Darwin V.5.158 [13] were used to analyze the collected data. Analyses of variance were performed with GenStat v9.1. For all these traits, genetic parameters were estimated from the components of the analysis of variance. Genotypic and Phenotypic Variances (VG and VP), Genotypic and Phenotypic Coefficients of Variation (GCV and PCV), heritability in the broad sense (H2) and expected Genetic Gain (GA) were calculated according to the formulas used by [14-18] presented in Table 2.
| Parameters | Formulas | Meaning of terms |
|---|---|---|
| Genotypic Variance (GV) | VG= (MSG-MSE)/r | MSG: Mean Square of Genotypes MSE: Mean Square Error r: Number of replicates |
| Phenotypic Variance (PV) | VP=VG+ (MSE/r) =MSG/r | |
| Heritability in the broad sense VG= (MSG-MSE)/r | H2 (%)= (VG/VP) × 100 | |
| Coefficient of Genotypic Variation (GCV) | GCV (%) = (√VG/X) × 100 | √VG: Standard deviation of genotypic variance √VP: Standard deviation of phenotypic variance I: Constant. with a selection of 5%, I is 2.06 X: trait mean |
| Coefficient of Phenotypic Variation (PCV) | PCV (%) = (√VP/X) × 100 | |
| Expected Genetic Gain (GA) | GA = H2 × √VP × I | |
| Expected genetic gain GA (% average trait) | GA (% average character) = (GA/X) × 100 |
Table 2: Formulas for estimated genetic parameters.
Variation in agro-morphological traits
Analysis of variance (Table 2) shows that fourteen characters discriminate between the ecotypes of Niger nutsedge, namely: Length of first leaf blade (LPL), length of second leaf blade (LDL), length of third leaf blade (LTL), width of third leaf blade (LaTL), diameter of mother plant (DPM), plant height (HP), number of tubers per bunch (NTP), tuber emission length (TEL), maximum root length (LMR), distance between mother plant and tillers (DPmT), number of flowering shoots (NHF), number of spikelets (NbrE), tuber yield (RT) and biomass yield (RB). Coefficients of variation for all characters ranged from 17.99% to 98.85%, reflecting strong variation between ecotypes. Plants of Niger nutsedge ecotypes vary in height from 30 cm to 90 cm, with low floral branching (5 to 14). Analysis of the table also shows that average tuber and above-ground biomass yields are 7.383 t/ha and 4.74 t/ha respectively as shown in Table 3.
| Characters | Minimum | Maximum | Moyenne (X) | MSG | MSE | CV (%) |
|---|---|---|---|---|---|---|
| LPL | 1.83 | 5.93 | 3.273 | 3.201* | 0.876 | 34.35 |
| LDL | 3 | 10.63 | 4.81 | 5.762* | 2.251 | 35.01 |
| LTL | 10.67 | 29.07 | 21.528 | 70.010** | 10.23 | 20.87 |
| LaPL | 0.1 | 0.6 | 0.143 | 0.011ns | 0.007 | 71.54 |
| LarDL | 0.11 | 0.5 | 0.203 | 0.011ns | 0.007 | 42.78 |
| LaTL | 0.2 | 0.79 | 0.507 | 0.086*** | 0.008 | 28.73 |
| DPM | 0.2 | 1.4 | 0.603 | 4.321*** | 0.028 | 51.59 |
| HP | 30 | 90 | 57.436 | 5963.160*** | 61.03 | 21.74 |
| NTP | 22 | 750 | 300.9 | 1023208.000*** | 11343 | 81.07 |
| LET | 3 | 52 | 20.644 | 4911.070*** | 13.36 | 75.88 |
| LMR | 10 | 55 | 28.638 | 1284.530*** | 40.76 | 27.21 |
| DPmT | 0.2 | 45 | 12.249 | 2447.650*** | 27.42 | 98.85 |
| NRF | 5 | 14 | 8.213 | 17.610*** | 1.757 | 17.99 |
| NbrE | 39 | 284 | 120.05 | 9465.000*** | 1450 | 33.99 |
| RdT | 0.868 | 13.877 | 7.383 | 51.248*** | 4.249 | 47.08 |
| RdB | 1.5 | 9 | 4.74 | 6.165* | 2.02 | 34.74 |
| Note: The length of the first leaf blade (LPL); the length of the second leaf blade (LDL); the length of the third leaf blade (LTL); the width of the first leaf blade (LarPL); the width of the second leaf blade (LaDL); the width of the third leaf blade (LaTL); the Diameter of the mother plant (DPM); the height of the plant (NP); number of tubers per bunch (NTP); tuber emission length (TEL); Maximum Root Length (MRL); distance between mother plant and tillers (DPmT); number of flowering stems (NHF); number of spikelets (NbrE); tuber yield (RdT) and biomass yield (RdB); *: Significant difference at 5%; **difference significant at 1%; ns: Not significant; ***: Highly significant: Mean square of genotype; MSE: Mean Square of Error; CV: Coefficient of variation. | ||||||
Table 3: Analysis of variance of the sixteen quantitative characters of nutsedge ecotypes.
Structuring the agromorphological variability of Niger nutsedge ecotypes
The results of the principal component analysis with all the measured traits reveal that the first five components explain 83.234% of the total variability (Table 4), including 44.769% for axis 1; 17.176% for axis 2; 12.188% for axis 3; and 9.102% for axis 4 (Table 4). Total variability is explained by 16 principal components with eigenvalues ranging from 0.734 to 7.163. The first axis is defined by growth-related traits (LTL, LaTL, DPmT, DPM, HP, LET) and yield (RdT and RdB). This component represents good vegetative development and high yield. The second component, which correlates with blade lengths (LPL and LDL), is that of height. The third axis, which contrasts Maximum Root Length (MRL) with flowering parameters (NHF and NbrE), is that of earliness. The fourth axis, which correlates with blade widths (LaPL and LaDL), is that of height, like the second axis.
| Principal components | F1 | F2 | F3 | F4 |
|---|---|---|---|---|
| Eigen values | 7.163 | 2.748 | 1.95 | 1.456 |
| Total variance (%) | 44.769 | 17.176 | 12.188 | 9.102 |
| Cumulative total variance (%) | 44.769 | 61.945 | 74.132 | 83.234 |
| LPL | 0.533 | 0.667* | 0.326 | -0.244 |
| LDL | 0.491 | 0.686* | 0.357 | -0.252 |
| LTL | 0.778* | 0.444 | 0.155 | -0.26 |
| LarPL | 0.386 | 0.263 | 0.204 | 0.808* |
| LaDL | 0.531 | 0.332 | 0.362 | 0.609* |
| LaTL | -0.753* | 0.418 | -0.1 | -0.269 |
| DPmT | -0.893* | 0.238 | -0.118 | 0.227 |
| DPM | -0.852* | 0.421 | 0.125 | 0.046 |
| HP | 0.890* | -0.146 | -0.114 | -0.051 |
| NTP | 0.59 | -0.58 | 0.027 | 0.147 |
| LET | -0.893* | 0.284 | -0.155 | 0.176 |
| LMR | -0.406 | 0.057 | 0.741* | -0.124 |
| NHF | 0.307 | 0.459 | -0.650* | 0.023 |
| NbrE | 0.367 | 0.523 | -0.666* | 0.072 |
| RdT | 0.821* | -0.236 | -0.008 | -0.187 |
| RdB | 0.709* | 0.255 | -0.165 | 0.066 |
| Note: The length of the first leaf blade (LPL); the length of the second leaf blade (LDL); the length of the third leaf blade (LTL); the width of the first leaf blade (LarPL); the width of the second leaf blade (LaDL); the width of the third leaf blade (LaTL); the Diameter of the mother plant (DPM); the height of the plant (NP); number of tubers per bunch (NTP); tuber emission length (TEL); Maximum Root Length (MRL); distance between mother plant and tillers (DPmT); number of flowering stems (NHF); number of spikelets (NbrE); tuber yield (RdT) and biomass yield (RdB); *: Significant difference at 5%; **difference significant at 1%; NS: Not significant; ***: Highly significant: Mean square of genotype; MSE: Mean Square of Error; CV: Coefficient of Variation; *: Correlated with axis (>0.6). | ||||
Table 4: Eigenvalues and percentage of variation expressed for the first five axes from the sixteen quantitative traits in principal component analysis.
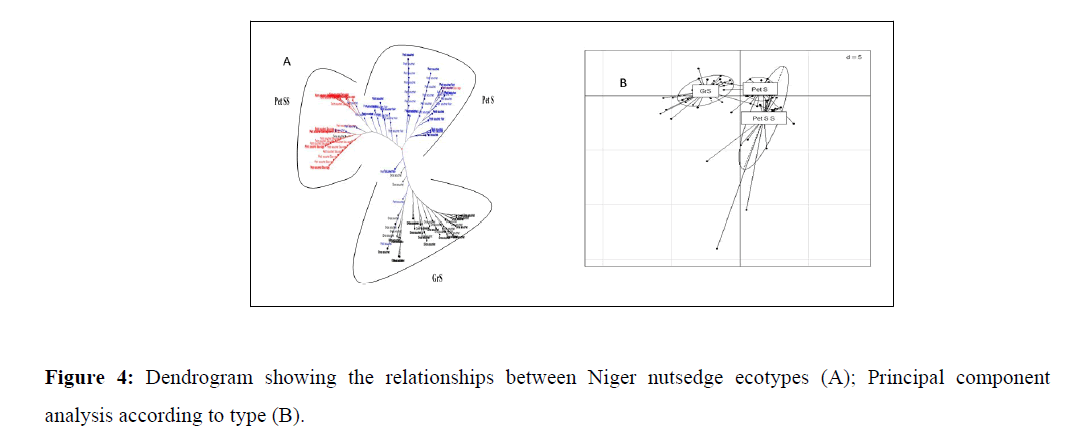
The dendrogram and the Principal Component Analysis (PCA) (Figure 4) shows a structuring of the individuals in the entire collection on a national scale. The general typology of the tree, grouping all the individuals, enables them to be divided into three (3) groups. The results of the Analysis of Variance (ANOVA) reported in Table 5 show highly significant differences at the 1% level between groups for the characters Length of Third leaf blade (LTL), width of third leaf blade (LaTL), diameter of mother plant (DPM), plant height (HP), Number of Tubers Per bunch (NTP), Tuber Emission Length (TEL), maximum root length (LMR), distance between mother plant and tillers (DPmT), number of flowering stems (NHF), number of spikelets (NbrE) and tuber yield (RdT). These results reflect morphological diversity between groups. Plant height (HP), Tuber Emission Length (TEL), distance between mother plant and tillers (DPmT), mother plant diameter (DPM) and tuber yield (RdT) were the main factors used to discriminate between groups. Thus, ecotypes in the petSS group have a fairly long tuber emission length (30.2 cm), mother plant and tillers distance (16.66 cm) and mother plant diameter (1.026), low plant height (39.8 cm) and low tuber yield (1, 768 t/ha), whereas ecotypes from the GrS and petS groups, with low tuber emission lengths (10.5 cm and 7.507 cm), mother plant and tillers distances (5.473 cm and 3.038 cm) and mother plant diameters (0.798 cm and 0.397 cm), recorded higher plant heights (56.08 cm and 63.76 cm) and yields (7.79 t/ha and 9.119 t/ha). Tuber yields of ecotypes in all groups increased as a function of plant height growth, tuber emission length and distance between mother plant and tillers.
| Characters | Collection | Groups | Probability | ||||
|---|---|---|---|---|---|---|---|
| Averages | Minimum | Maximum | GrS | petS | petSS | P (value) | |
| LPL | 3.273 | 1.83 | 5.93 | 3.758 | 3.407 | 2.386 | 0.022 |
| LDL | 4.81 | 3 | 10.63 | 5.71 | 4.925 | 3.566 | 0.07 |
| LTL | 21.528 | 10.67 | 29.07 | 23.61 | 22.925 | 15.26 | 0.001 |
| LaPL | 0.1428 | 0.1 | 0.6 | 0.146 | 0.153 | 0.108 | 0.646 |
| LaDL | 0.2032 | 0.11 | 0.5 | 0.222 | 0.216 | 0.146 | 0.226 |
| LarTL | 0.5068 | 0.2 | 0.79 | 0.532 | 0.428 | 0.718 | 0.001 |
| PM | 0.6028 | 0.2 | 1.4 | 0.798 | 0.397 | 1.026 | 0.001 |
| HP | 57.436 | 30 | 90 | 56.08 | 63.767 | 39.8 | <0.001 |
| NTP | 204.9 | 22 | 750 | 62.18 | 298.1 | 68.08 | 0.001 |
| LET | 12.644 | 3 | 52 | 10.5 | 7.507 | 30.2 | <0.001 |
| LMR | 28.638 | 10 | 55 | 35.12 | 25.957 | 30.2 | 0.001 |
| DPmT | 6.249 | 0.2 | 45 | 5.473 | 3.038 | 16.66 | 0.001 |
| NHF | 8.213 | 5 | 14 | 7.867 | 8.378 | 8.067 | 0.001 |
| NbrE | 120.05 | 39 | 284 | 105.8 | 125.233 | 118.77 | 0.001 |
| RdT | 7.383 | 0.868 | 13.877 | 7.79 | 9.119 | 1.768 | <0.001 |
| RdB | 4.74 | 1.5 | 9 | 4.5 | 5.367 | 3.1 | 0.041 |
| Note: The length of the first leaf blade (LPL); the length of the second leaf blade (LDL); the length of the third leaf blade (LTL); the width of the first leaf blade (LarPL); the width of the second leaf blade (LaDL); the width of the third leaf blade (LaTL); the Diameter of the mother plant (DPM); the height of the plant (NP); number of tubers per bunch (NTP); tuber emission length (TEL); Maximum Root Length (MRL); distance between mother plant and tillers (DPmT); number of flowering stems (NHF); number of spikelets (NbrE); tuber yield (RdT) and biomass yield (RdB); *: Significant difference at 5%; **difference significant at 1%; NS: Not significant; ***: Highly significant: Mean square of genotype; MSE: Mean Square of Error; CV: Coefficient of variation; GrS: Big nutsedge; petS: Small nutsedge; petSS: Small wild nutsedge; Pr: Probability of F. | |||||||
Table 5: Average performance of the three groups of Niger nutsedge for discriminating characteristics.
Estimation of genetic parameters
Phenotypic and genotypic variances: The genetic parameter data in Table 6 show that the phenotypic variance is greater than the genotypic variance for all the different variables or parameters studied. Phenotypic variance ranges from 0.002 to 20464.160 and genotypic variance from 0.001 to 20237.300. Number of tubers (20464.160) showed the highest phenotypic variance, followed by number of spikelets (315.500), plant height (119.263), tuber emulsion length (98.221), distance between mother plant and tillers (34.966) and maximum root length (25.691). The length of the third blade shows phenotypic and genotypic variances. Moderate (<20) respectively of 14.002 and 11.956 respectively. The other traits had low genotypic and phenotypic variances (<10), with the exception of tuber yield, which had a moderate phenotypic variance of 10.250.
| Characters | VG | VP | H2 (%) | √VG | √VP | GCV (%) | PCV (%) | GA | GA (% average character) |
|---|---|---|---|---|---|---|---|---|---|
| LPL | 0.465 | 0.64 | 72.633 | 0.682 | 0.8 | 20.835 | 24.447 | 1.197 | 36.579 |
| LDL | 0.702 | 1.152 | 60.934 | 0.838 | 1.073 | 17.421 | 22.318 | 1.347 | 28.014 |
| LTL | 11.956 | 14.002 | 85.388 | 3.458 | 3.742 | 16.062 | 17.382 | 6.582 | 30.574 |
| LaPL> | 0.001 | 0.002 | 36.767 | 0.029 | 0.047 | 20.151 | 33.232 | 0.036 | 25.17 |
| LaDL | 0.001 | 0.002 | 35.544 | 0.028 | 0.046 | 13.596 | 22.804 | 0.034 | 16.697 |
| LaTL | 0.016 | 0.017 | 90.54 | 0.125 | 0.131 | 24.673 | 25.93 | 0.245 | 48.363 |
| DPM | 0.086 | 0.086 | 99.358 | 0.293 | 0.294 | 48.611 | 48.767 | 0.602 | 99.816 |
| HP | 118.043 | 119.263 | 98.977 | 10.865 | 10.921 | 18.916 | 19.014 | 22.267 | 38.768 |
| NTP | 20237.3 | 20464.2 | 98.891 | 142.258 | 143.053 | 47.277 | 47.542 | 29.142 | 96.85 |
| LET | 97.954 | 98.221 | 99.728 | 9.897 | 9.911 | 47.942 | 48.008 | 20.36 | 98.626 |
| LMR | 24.875 | 25.691 | 96.827 | 4.988 | 5.069 | 17.416 | 17.699 | 10.11 | 35.303 |
| DPmT | 34.575 | 34.966 | 98.88 | 5.88 | 5.913 | 48.004 | 48.275 | 12.045 | 98.333 |
| NHF | 0.528 | 0.587 | 90.023 | 0.727 | 0.766 | 8.851 | 9.329 | 1.421 | 17.3 |
| NbrE | 267.167 | 315.5 | 84.68 | 16.345 | 17.762 | 13.615 | 14.796 | 30.985 | 25.81 |
| RdT | 9.4 | 10.25 | 91.709 | 3.066 | 3.201 | 41.527 | 43.363 | 6.048 | 81.922 |
| RdB | 0.829 | 1.233 | 67.234 | 0.91 | 1.11 | 19.209 | 23.426 | 1.538 | 32.446 |
| Note: The length of the first leaf blade (LPL); the length of the second leaf blade (LDL); the length of the third leaf blade (LTL); the width of the first leaf blade (LarPL); the width of the second leaf blade (LaDL); the width of the third leaf blade (LaTL); the Diameter of the mother plant (DPM); the height of the plant (NP); number of tubers per bunch (NTP); tuber emission length (TEL); maximum root length (MRL); distance between mother plant and tillers (DPmT); number of flowering stems (NHF); number of spikelets (NbrE); tuber yield (RdT) and biomass yield (RdB); *: Significant difference at 5%; **difference significant at 1%; NS: Not significant; MSE: Mean Square of Error; CV: Coefficient of Variation; VG: Genotypic variance; VP: Phenotypic variance; H2 : Heritability in the broad sense; GCV: Coefficient of genotypic variation; PCV: Coefficient of phenotypic variation; √VG: standard deviation of genotypic variance; √VP: standard deviation of phenotypic variance; GA: Expected genetic gain; GA (% trait mean); genetic gain relative to the trait mean. | |||||||||
Table 6: Calculated genetic parameters of Niger nutsedge ecotypes.
Phenotypic and genotypic coefficients of variation: For all traits studied, phenotypic coefficients of variation are higher than genotypic coefficients of variation. According to [19] cited by [20] the coefficients of genotypic and phenotypic variation are low below 11%, moderate between 11% and 20% and high above 20%. Thus, low phenotypic and genotypic coefficients of variation are observed for the characters number of floral branches (8.851% and 9.329%). The vegetative characters, namely second blade length (17.421 and 22.31), third blade length (16.062 and 17.382), second blade width (13.596 and 22.804), maximum root length (17.416 and 17.699), number of spikelets (19.209 and 23.426) and biomass yield (19.209 and 23.426) have moderate genotypic and phenotypic coefficients of variation, while the other characters have high coefficients of variation.
Heritability in the broad sense: According to Johnson (1955) and Stanfield (1975), heritability is high above 50%, low below 20% and average between 20% and 50%. Heritability ranged from 35.544% to 99.358% for all the traits studied, except for the width of the first and second leaf blades (36.767% and 35.544%), which showed medium heritability.
Expected genetic gain: Expected genetic gain relative to the trait mean (GA (% trait mean)) ranged from 16.697% for second blade width to 99.816% for mother plant diameter. Genetic gain was high for all traits except number of flowering stems (NHF) (17.300%) and second blade width (LaDL) (16.697%). Traits such as diameter of mother plant (DPM), Number of Tubers per Plant (NTP), Tuber Emission Length (TEL), distance between mother plant and tillers (DPMT) and tuber yield (RdT) recorded the highest genetic gains (>80%) (Table 6).
Discussion
The analysis of variance carried out on the quantitative characteristics of nutsedge ecotypes shows significant differences between minimum and maximum values, as well as high coefficients of variation for these characteristics, reflecting the existence of considerable morphological variability within ecotypes. Similar observations were also made by [21] on 64 accessions and [22] on 35 accessions. In their study on the valorization of four nutsedge (Cyperus esculentus L.) accessions from Cameroon, [23] affirm that tiger nut accessions show genetic diversification within the populations tested in terms of growth and yield parameters. These results are also corroborated by those of [24] who asserted that the populations of tiger nut (Cyperus esculentus L.) possessed the greatest genetic diversity populations of the neighboring species Cyperus rodondus. The ecotypes of both groups (cultivated tiger nut) resulting from the hierarchical ascending classification are observed in all the regions surveyed, proof of the maintenance of diversity in situ by farmers. This could be linked to seed exchanges between local communities. This structuring is also comparable to that of the Belgian Tiger nut ecotypes studied by [22] on a sample of 35 accessions, which also identified three morphological groups. The Niger and Belgian nutsedge ecotypes would be less diverse than those of the nutsedge studied by [25-27] where they respectively obtained 4 morphological groups on Burkina nutsedge, 5 morphological groups on 42 accessions of nutsedge from the temperate zone of China and 6 groups on 34 accessions of nutsedge from Ghana. The different morphological groups obtained offer a possible choice of genitors for the creation of new varieties to meet growers' needs [18]. If the aim of selection is yield, it could be directed towards group 1 and 2 ecotypes, which have the best tuber yields per plant. But if the selection objective is nutritional, the choice may be directed towards group 3, whose ecotypes are very rich in iron (15.809 ± 6.524 mg/100 g) [12]. The estimation of genetic parameters showed that the genotypic variance is lower than the phenotypic variance for all traits. These results are in line with those of [20] on sweet sorghum, those of [28] on millet, [17] on rice and [29] on mulberry cited by [20]. The study showed little difference between the values of the two variances for all traits [20]. Asserted that this state of affairs would indicate a weak influence of the environment on the expression of the traits and a greater genetic control over their expression. This weak influence of the environment on tiger nut ecotypes is also confirmed by the high heritability obtained by almost all the traits studied. According to [19-30], "heritability alone does not predict whether selection will bring substantial improvement. However, the joint estimation of heritability and expected genetic gain (GA (% mean of trait)) can provide more reliable information". The characteristics of the nutsedge ecotypes studied showed high to very high heritability in the broad sense and expected genetic gain, indicating additive gene action [14-31]. With the exception of the width of the first leaf blade (LaPL), the width of the second leaf blade (LaDL) and the number of flowering stems (NHF), which showed non-additive gene action.
Conclusion
The study showed the existence of very wide agro-morphological variability within the ecotypes of Niger nutsedge, organised around fourteen vegetative traits and yield. A structuring of the ecotypes into three morphological groups was obtained on the basis of: Plant height (HP), Tuber Emission Length (TEL), distance between mother plant and tillers (DPmT), mother plant diameter (DPM) and tuber yield (RdT). The PetS group performed well in terms of tuber production per bunch and tuber yield. Estimation of the genetic parameters also showed that environmental factors had little influence on the expression of the traits, which was reflected in the small differences between the phenotypic and genotypic coefficients of variation. High to very high heritability in the broad sense, coupled with high expected genetic gain, were observed for most of the traits studied, showing that genes with additive effects are involved in the expression of these traits. Improvement by direct selection is therefore possible for these traits.
Acknowledgments
The authors acknowledge and would like to express their gratitude to the Researchers for National Institute of Agronomic Research of Niger, Plant Production and Biodiversity Department of Environment Science Faculty, University of Diffa (Niger) and Laboratory for the Management and Valorization of Biodiversity in the Sahel (GeVaBioS)/UAM for supporting the study and during part of this work.
Conflicts of Interest
We, the authors, declare that there are no conflict of interest in this manuscript.
References
- De Castro O, Gargiulo R, Guacchio ED, Caputo P, de Luca P (2015) A molecular survey concerning the origin of Cyperus esculentus (Cyperaceae, Poales): Two sides of the same coin (weed vs. crop). Ann Bot 115: 733-745.
[Crossref], [Google Scholar], [Indexed]
- Ban-Koffi L, Nemlin GJ, Le Fevre S, Kamenan A (2005) Physico-chemical characterisation and therapeutic potential of sweet pea (Cyperus esculentus L., Cyperaceae). Agron Afr 17: 63-71.
[Crossref], [Google Scholar]
- Gado I, Oumarou C, Mahaman H, Karimou I, Ibrahim B (2004) Effet de la fertilisation minérale et organique sur le gros souchet. INRAN/PPEAP Niamey Niger 8p.
- Toukoua D, Sidikou D, Ibrahim A, Badage G (2002) Plan d'action sur la filière souchet. Ministère de l'agriculture et de l'élevage. Niamey/Niger 99p.
- Daouda A (2008) Contribution à l'étude de la culture du souchet (Cyperus esculentus L.) dans le village d'Atchidakofato. FA/UAM. Niamey 40p.
- Dodet M (2006) Diversité génétique et phénologie de Cyperus esculentus L. (Cyperaceae) pour une gestion intégrée de l'espace dans les cultures de Haute Lande. Theses 226p.
- Schippers P, Ter Bog SJ, Bos JJ (1995) A revision of the infraspecific taxonomy of Cyperus esculentus (yellow nutsedge) with an experimentally evaluated character set. Syst Bot 20: 461-481.
- Kazan K, Manners JM, Cameron DF (1993) Genetic variation in agronomically important species of Stylosanthes detemined using random amplified polymorphic DNA markers. Theor Appl Genet 85: 882-888.
[Crossref], [Google Scholar], [Indexed]
- Chebbil H, Pascual-Villalobos MJ, Cenis JL, Correal E (1995) Morphological and molecular characterisation of woody species of the genus Medicago. CABI Databases 142: 191-206.
- Douhovnikoff V, Dodd RS (2003) Intra-clonal variation and a similarity threshold for identification of clones: Application to Salix exigua using AFLP molecular markers. Theor Appl Genet 106: 1307-1315.
[Crossref], [Google Scholar], [Indexed]
- Haoua B, Halima OD, Iro DG, Toudou A (2019) Nutritional and mineral compositions of nutsedge (Cyperus esculentus L.) tubers from different ecotypes in Niger. Asian J Crop Sci 8: 39-48.
[Crossref]
- Haoua B, Sani IS, Hamissou Z, Toudou A (2023) Study of the genetic diversity of tiger nut (Cyperus esculentus) ecotypes of Niger using SSR markers. International Journal of Genetics and Genomics 11: 18-26.
- Perrier X, Jacquemoud-Collet JP (2006) Darwin software.
- Johnson HW, Robinson HF, Comstock RE (1955) Estimates of genetic and environmental variability in soybeans. J Agron 47: 314-318.
[Crossref], [Google Scholar]
- Assefa K, Tefera H, Merker A, Kefyalew T, Hundera F (2001) Variability, heritability and genetic advance in phenomorphic and agronomic traits of tef (Eragrostis tef (Zucc.) Trotter) germplasm from eight regions of Ethiopia. Hereditas 134: 103-113.
[Crossref], [Google Scholar], [Indexed]
- Rex B (2002) Breeding for quantitative traits in plants. Stemma press, Woodbury, Minnesota, USA, p: 369.
- Hosseini SJ, Sarvestani ZT, Pirdashti H, Afkhami A, Hazrati S (2012) Estimation of heritability and genetic advance for screening some rice genotypes at salt stress conditions. Intl J Agron Plant Prod 3: 475-482.
- Sawadogo N, Nebié B, Kiébré M, Bationo-Kando P, Nanema KR, et al. (2014) Agromorphological characterization of sweet grain sorghums (Sorghum bicolor (L.) Moench) from Burkina Faso. Int J Biol Chem Sci 8: 2183-2197.
[Crossref], [Google Scholar]
- Govindaraj M, Selvi B, Rajarathinam S, Sumathi P (2011) Genetic variability and heritability of grain yield components and grain mineral concentration in India's pearl millet (Pennisetum glaucum (L) R. Br.) accessions. Afr J Agric Res 5: 3060-3064.
[Crossref], [Google Scholar]
- Sawadogo N, Nanema KR, Bationo P, Traore RE, Nebie B, et al. (2014) Evaluation of the genetic diversity of sweet grain sorghums (Sorghum bicolor (L.) Moench) from northern Burkina Faso. J Appl Biosci 84: 7654-7664.
[Crossref], [Google Scholar]
- Asare PA, Kpankpari R, Adu MO, Afutu E, Adewumi AS (2020) Phenotypic characterization of tiger nuts (Cyperus esculentus L.) from major growing areas in Ghana. Sci World J 2020: 7232591.
[Crossref], [Google Scholar], [Indexed]
- de Ryck S, Reheul D, de Riek J, de Keyser E, de Cauwer B (2023) Genetic and morphological variation of belgian Cyperus esculentus L. clonal populations and their significance for integrated management. Agronomy 13: 572.
[Crossref], [Google Scholar], [Indexed]
- Sakatai DP, Bouba AA, Bassala JPO, Balna J, Palou O, et al. (2020) Valorization of four accessions of nutsedge (Cyperus esculentus L.): A good crop diversification option for producers in the locality of danay-vokgora (Mayo-danay) in the Sudano-Sahelian zone of Cameroon. Int J Biol Chem Sci 14: 2277-2293.
[Crossref], [Google Scholar]
- Tayyar RI, Nguyen JHT, Holt JS (2003) Genetic and morphological analysis of two novel tiger nut biotypes from California. Weed Science 51: 731-739.
[Crossref], [Google Scholar]
- Yamwembo S, Ouedraogo J, Sawadogo P, Sawadogo N, Ouedraogo H, et al. (2022) Agronomic variability of yellow nutsedge (Cyperus esculentus L.) accessions in Burkina Faso. Int J Innov 9: 1-11.
- Yang X, Niu L, Zhang Y, Ren W, Yang C, et al. (2022) Morpho-agronomic and biochemical characterization of accessions of tiger nut (Cyperus esculentus) grown in the North temperate zone of China. Plants (Basel) 11: 923.
[Crossref], [Google Scholar], [Indexed]
- Donkor E, Nyadanu D, Dapaah H (2019) Germplasm collection and morphological characterization of local accessions of tiger nut (Cyperus esculentus L.) in Ghana for conservation and utilization. J Plant Breed Crop Sci 11: 196-205.
[Crossref], [Google Scholar]
- Drabo I, Zangre RG, Sawadogo M, Ouedraogo M (2013) Genetic variability and estimates of genetic parameters in Burkina Faso's pearl millet landraces. Int J Agric Sci 3: 367-373.
[Crossref], [Google Scholar]
- Doss SG, Chakraborti SP, Roychowdhuri S, Das NK, Vijayan K, et al. (2012) Variability, heritability and genetic advance in mulberry (Morus spp) for growth and yield attributes. Agricultural Sciences 3: 208-213.
[Crossref], [Google Scholar]
- Govindaraj M, Selvi B, Kumar IS (2011) Genetic diversity studies in Indigenous pearl millet (Pennisetum glauccum (L.) R. Br.) accessions based on biometrical and nutritional quality traits. Indian J Plant Genet Resour 24: 186-193.
- Kashif M, Ahmad J, Chowdhry MA, Perveen K (2003) Study of genetic architecture of some important agronomic traits in durum wheat (Triticum durum Desf.). Asian J Plant Sci 2: 708-712.
[Crossref]

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences
 : Communaute urbaine de Niamey;
: Communaute urbaine de Niamey;  : Capitale du Niger;
: Capitale du Niger; : Site
d’experimentation agro-morphologique et moleculaire
: Site
d’experimentation agro-morphologique et moleculaire