ISSN : 2249 - 7412
Asian Journal of Plant Science & Research
Elucidation of Musa acuminata cv. Berangan Root Infection by FOC (Tropical Race 4) by RNA Sequencing and Analysis
Umaiyal Munusamy1*, Kamilatulhusna Zaidi2,3, Nadiya Akmal Baharum2,3,4, Ho Hui Li2,3, Yusmin Mohd-Yusuf2,5, Rofina Yasmin Othman2,3*
1Institute of Plantation Studies, Universiti Putra Malaysia, Selangor, Malaysia
2Centre for Research in Biotechnology for Agriculture (CEBAR), University of Malaya, Kuala Lumpur, Malaysia
3Institute of Biological Sciences, University of Malaya, Kuala Lumpur, Malaysia
4Department of Cell and Molecular Biology, Universiti Putra Malaysia, Selangor, Malaysia
5Centre for Foundation Studies in Science, University of Malaya, Kuala Lumpur, Malaysia
Abstract
Musa acuminata cv. Berangan (AAA) is a type of banana locally grown in Malaysia. These bananas as well as Musa acuminata cv. Cavendish (AAA) are also facing a major threat from a typical soil borne fungus identified as Fusarium oxysporum f. sp. cubense race 4 (FocR4). Its characteristics as a complex pathogen manifesting as subtypes or races are the main reasons it is difficult to control its infections. Genome sequence availability of the double haploid Musa acuminata originating from Pahang has become very useful to analyse RNA-seq reads and also to identify the transcriptome profile of the host response between different groups. High throughput sequencing was accomplished using RNA-Seq technology based on the Illumina HiSeqTm 2000 platform. Three sets of libraries derived from infected and mock infected plants (experimental groups) between different time points (0, 48, 96 h) shows over forty million reads were generated, each corresponding to coverage of >4,000,000,000 to <8,000,000,000 bases. About 0.10-66% reads were mapped to Musa acuminata DH Pahang genome sequence. This study provides the statistical analysis of the sequence reads. Based on this information, further analysis on gene expression patterns influenced by Foc race infection within the tested groups and time points will help in the understanding of the host pathogenic responses. In future, excessive transcriptomic data will promise a discovery of many new genes for plant infection diagnosis.
Keywords
Musa acuminata cv. ‘Berangan, root infection; FOC (Tropical Race 4); RNA sequencing
Introduction
Musa acuminata is a popular banana produced on a large scale around Asia and Africa. In Asia, India is the largest producer followed by Uganda, China and Philippines [1]. Although Musa acuminata variety Cavendish is exported globally, cultivation of different varieties of banana in different regions for local consumption are varied. Musa acuminata cv. Berangan is native to the Asian tropics such as Malaysia, Indonesia, Philippines, Australia, and also East Africa. It is a popular cultivar consumed as a dessert [2,3].Berangan has similar properties with Cavendish such as it consists of an acceptable level of acidity, has a slightly dry starchy texture, good flavour and a reasonable shelf life compared to the rest of the other local varieties [4].
However, this cultivar is also under attack by similar wilting diseases as in Cavendish. The wilting disease that is caused by Fusarium oxysporum f. sp. cubense (Foc) is a type of soil borne [5]. It is a complex pathogen that manifested as subtypes or races. They are responsible for the outbreaks in Latin America, The Carribean Islands, Taiwan, The Phillipines, Malaysia, Indonesia, The Northern Territory of Australia and China [6-11]. Furthermore, the most virulent fusarium strain identified as Foc Tropical Race 4 (FocR4) is currently not limited in tropical regions in Asia but has alarmingly diagnosed in Mozambique and Jordan and most recently was diagnosed in Northern Queensland [8]. This indicates that Foc Tropical Race 4 is emerging into broader regions and both cultivars might be destroyed by Fusarium wilt [12]. Therefore, in-depth knowledge and information on the interactive response information are important in developing strategies to identify and to overcome these emerging diseases.
Advancement in transcriptome approaches are being utilized widely to conduct investigation on gene expression in response to fusarium wilt infection in bananas. Furthermore, the breakthrough of Musa acuminata and Musa balbisiana genomes have made ease in designing, collecting and analysing gene expression data have highlighted differentially expressed genes in bacterial pathogenesis in bananas using transcriptome approach [13]. Following that, have reported a new approach to study differentially expressed genes by inducing a biochemical process through herbivory infection in banana plants [14]. By applying a transcriptome approach, more detailed data on the biochemical process was carried out successfully. With this unlimited information, the complexities of the disease infection network can be accurately identified [15,16].
Based on reported studies. FOC pathogenesis studies have mainly focused on Musa acuminata cv. Cavendish, however less work was reported based on regional cultivars such as Berangan [17-22]. Plant researchers utilize a variety of approaches to understand gene expression in many banana varieties. In some cases, transcriptome profiling based on reference guided is preferred than reference independent transcriptome profiling. In order to study plant functional genomics, next generation sequencing technologies enable plant researchers to perform studies in any plant species with higher dynamic range with lower cost compared to traditional microarray technology which can only be used for gene expression profiling in species with known transcriptome sequences.
This study analysed the transcriptome responses on early infection of infected Musa acuminata cv. Berangan in the greenhouse (pre-field screening). This pre-field screening assay will be an important tool to control and compare gene expression analysis that can be utilized to further understand the host response towards infection and identify early infection responses.
Materials and Methods
Plant Material and Fusarium oxysporum Race 4 (FocR4)-C1 HIR
Tissue culture-derived banana Musa acuminata cv. ‘Berangan’ plantlets were obtained from CEBAR University Malaya, Malaysia. Plantlets were maintained in Murashige and Skoog (MS) medium for 1 month. For rooting, MS active charcoal (10 g/L) was used [23]. Healthy plantlets aged 2 months with at least 3-5 green leaves with a minimum length of 5 cm of white roots, and stem diameters of between 0.5 to 1.0 cm were chosen for the infection studies. Isolate C1 HIR Fusarium oxysporum Race 4 (FocR4) which wgoas maintained as pure culture on water agar using gelrite (Duchefa Biochemie, Netherlands) at the PhytoMycology Laboratory, University Malaya were used.
Sample Collection
Infected root samples from 2, 48 and 96 h were sampled. The infected plant roots were uprooted and washed with distilled water and immediately stored in liquid nitrogen. Each biological replicate from different time points were labeled separately and stored in -80ºC for subsequent analysis.
RNA Extraction and Quality Control
Approximately 0.5g of the harvested roots were grinded for total RNA extraction using RNeasy® Plant Mini Kits, (Qiagen, Germany). The RNA extraction protocol was followed by the manufacturer’s instructions. The purity of total RNA was determined by NanoDrop ND-1000 spectrophotometer (NanoDrop, USA) AT 260/280 NM ratio 2.0 and 260/230 (ratio 2.2). Total RNA concentration and integrity were determined using an Agilent 2100 Bioanalyzer with a minimum integrity number at least 8 and 1% gel electrophoresis analyses, respectively.
mRNA Purification
mRNA purification kit was used to pool mRNA with a poly-A tail. The purified RNA was randomly fragmented and was reverse transcribed into cDNA. Adapters were ligated into this fragment prior to PCR. Fragments with 200- 400 bp length were selected for paired end sequencing.
Library Preparation
The mRNA content was recovered from total RNA as described in Illumina TruSeq RNA library Prep Kit. The mRNA was captured twice on poly-T oligo magnetic beads prior to fragmentation using a fragmentation buffer. The fragmented strand was used to synthesize the first cDNA strand by priming with random hexamers. The second strand was generated and was purified using Ampure XP beads (Illumina, UK). A single adenine base was added to the 3` ends and sequencing adaptors were then ligated to the fragments and a flow cell was used to select the range of fragments suitable for PCR amplification. The quality control analysis for the sample library and quantification of the DNA library template was performed prior to sequencing. Sequencing was carried out using an Illumina Hi Seq™ 2000 platform.
The quality control of the sequence reads were analyzed based on overall reads quality, total bases, total reads and GC content. Artifacts such as low quality reads, adapter sequence, contaminant DNA and PCR duplicates were removed. Aligned reads were generated using TopHat prior alignment against the reference genome. Transcript assembly of aligned reads were generated using cufflinks. Expressed profile was calculated based on mapped transcript per sample. Normalization of transcript length and depth coverage was carried out to compare expression profile between samples. Reads Per Kilobase of transcript per million mapped reads (RPKM) values were used in normalization.
RNA-Seq Quantification
RNA Seq quantification is a current tool for gene expression profiling that is based on next generation sequencing (NGS) technology. It can simultaneously interrogate tens of thousands of transcripts and provide precise measurement of their expression levels. Compared with microarray based methods, RNA Seq quantification provides greater sensitivity, accuracy and broader dynamic range. Therefore, RNA sequencing quantification is widely used in plant disease research [16,24-26]. The digital signal that comes with low background noise is an added advantage of this technique. It has a high accuracy, reproducibility, sensitivity and has a wide dynamic range for gene expression studies using the RPKM method.
Computing Resources, Data Processing, Quality Control and File Formats
CLC Genomic Workbench software was used to analyze the sequence reads. NCBI database was utilized (http://www.ncbi.nlm.nih.gov/genome/10976) to download genome sequences and gene annotation files. The reads were functionally annotated using Blast2Go software. Software such as Aspera, FileZilla and Blast2Go were downloaded to the available LINUX system. Cross-platform file formats including fasta, fastq, sam, bam, gtf and gff files were used. Raw data with adjunct sequences were processed before being mapped. Basic tasks such as adapter removal, trimming quality was set to 0.001 and the summary statistics on quality score was performed by Q30.
Direct Link to Deposited Data
Data was deposited at
‘https://www.ncbi.nlm.nih.gov/biosample?LinkName=bioproject_biosample_all&from_uid=287860’
Reproducibility of Transcriptome Profiles
The raw RNA-Seq reads were processed by FastQC (version: v0.10.0) to remove the low quality reads through a modular set of analyses and then was mapped to PKW pseudo-chromosome genome (transcriptome re-seq reference genome in http://banana-genome.cirad.fr) using a fast splice junction mapper Tophat (version: v1.3.3). This aligns the RNA Seq reads to the reference genome (PKW_pseudochromosome) through ultra high-throughput short read aligner Bowtie. The mapping results were analyzed to identify the splice junctions between exons. Transcript abundance of the novel gene and expression level of mapped genes were calculated with the program Cufflinks (version: v2.1.1). Gene expression levels represented as volcano plots were normalized with reads per kilobase of exon per million mapped reads (RPKM) values. Aligned RNA sequence reads were assembled into a parsimonious set of transcripts. Estimates of the relative abundance of these transcripts were based on the number of reads that support each one while taking into account the biases in library preparation protocols. Scatter plot, PCA plot and Box-Plot were also used to determine the reproducibility of the transcriptome profiles. In scatter plot, to examine the variability among RNA-seq experiments, all clean reads from infected samples for 48 and 96 h were displayed in scatter plot with 2 h infected sample as control. Infected samples of 48 and 96 h and control sets of 2 h were plotted for all possible pairs of independent experiments. To cluster the samples based on the similarity of gene expression profiles principal component analysis (PCA) were used. While in Box-plot representations of the up-regulated and down-regulated gene expressions in all 3 experimental infections. Paired t-test was used in statistical analysis. FPKMs, fragments per kilobase of exon per million fragments mapped.
Gene Functional Annotation and Classification
For functional analysis of the unigenes, Gene Ontology (GO) annotations were determined using the Blast2 GO program. The KEGG database (http://www.genome.jp/kegg/) was used to achieve pathway annotations and the KEGG mapper was used to identify DEGs that the pathways showed [27].
Pathway Assignment
To characterize the pathway enrichment of the identified DEGs, gene classification was performed on the basis of KEGG analysis. The GO number was obtained for each protein and was used for constructing metabolic pathways [28].
Transcriptomic Validation
To further validate the transcriptomic profile of RNA Seq, genes with increased and reduced expression were chosen for qPCR analysis. Various primers were designed for a particular gene and only primers that produced single fragments of the expected lengths were used in qPCR amplification analysis.
Sample Preparation
Total RNA was extracted with RNeasy® Plant Mini Kits from Qiagen, Germany according to the manufacturer’s instructions. DNA contamination was eliminated using DNase. RNA quality was evaluated using an Agilent 2100.
cDNA Library Preparation
The purified RNA was randomly fragmented and was reverse transcribed into cDNA. Adapters were ligated into this fragment prior to qPCR. Only 200- 400 bp fragment was selected for paired end sequencing.
Real-Time PCR (qPCR) Primers
To further confirm the validity of the transcriptome data real time assay was carried out. The real time expression profiles of banana defense-related genes were analysed in cDNA samples that were obtained from both infected and non-infected banana roots. Total of 23 genes were pooled through transcriptome data that was submitted in NCBI gene bank (NCBI SRA submission, Accession: PRJNA287860). Primers were designed by using Primer3 software. List of primer sequences were listed in Table 1 40S Ribosomal protein S2 (RPS2), were chosen as the housekeeping gene.
RT-qPCR Conditions
Real-time analysis was performed in an Applied Biosystem 7500 Fast Real Time System using KAFA SYBR FAST qPCR Kit Master Mix (2X) (Universal, United States). The reaction mixture consists of 1 μl of cDNA sample, 10μl of KAPA SYBR FAST qPCR Master Mix (2x) Universal, 0.4 μl of forward and reverse real time primers and 0.4 μl ROX low. Non-template reactions (NTC) containing nuclease free water were used. 20 μl of the mixtures were distributed evenly into MicroAMP™ Optical 8-Tube Strips (Applied Biosystem, USA). Amplification cycles were conducted as follows: Initial denaturation at 95°C for 10 min, thermal cycling was performed for 40 cycles with 92ºC for 15 sec and 60ºC for 120 sec with the fluorescence being read at the end of each cycle. Dissociation curve was analyzed at 95ºC followed by 60°C after each completed run to evaluate the presence of non-specific PCR products and primer dimers amplification.
Results and Discussion
Library Quality Control (QC) Result of RNA
Table 1 showed the quality control result of the extracted RNA. It has become the most widely used material in next generation sequencing technology [29]. The QC analysis which was carried out before sequencing verifies the expected insert size with no contamination of adapter-dimers [30]. Contamination introduced during the library preparation can generate sequencing errors during sequencing and base calling steps [31]. In addition, the importance to determine the quality control of RNA is because RNA-Seq technology has higher productivity and better resolution to generate mainstream of high throughput of large scale RNAs information such as measures the abundance and structure of genes at the RNA level, and employs different analytical approaches [29]. Therefore, only passed QC RNA are used for RNA sequencing.
| Library Name | Library Type | Concentration g/uL | Concentration (nM) | Size | Result |
|---|---|---|---|---|---|
| Zero 1 | Truseq RNA | 37.85 | 210.19 | 277 | Pass |
| D0 9889-6 | Truseq RNA | 82.88 | 445.84 | 286 | Pass |
| DAY0-RNA1b | Truseq RNA | 71.66 | 388.17 | 284 | Pass |
| D2-1 | Truseq RNA | 38.53 | 194.37 | 305 | Pass |
| D2 9889 5 | Truseq RNA | 87.99 | 480.06 | 282 | Pass |
| Day 2 9889 tube 2 | Truseq RNA | 63.8 | 333.87 | 294 | Pass |
| D4-3 | Truseq RNA | 28.83 | 150.88 | 294 | Pass |
| Day4 9889 G | Truseq RNA | 118.47 | 650.95 | 280 | Pass |
| DAY 4 9889 tube 2 | Truseq RNA | 56.42 | 305.64 | 284 | Pass |
Table 1: Library QC result of RNA.
Output Statistics of RNA Seq Libraries
Table 2 showed the output statistics of the raw transcriptome and reference mapping of Musa acuminate cv. Berangan infected with FocR4. It generated a total of 7 billion to 700 hundred million reads per library. After removal of reads including adaptor sequence, ambiguities which are limit to two nucleotides, filtered on length with 25 nucleotides short reads of the total number of base pairs sequence varied from 40 million to 77 million. Total trimmed reads ranged from 2 million up to 6 million while trimmed nucleotides varied from 800 million up to 1 billion. The proportion of clean reads with Q30 scores were exceeded ≥ 90% for all the sequenced samples. This demonstrated that the assay has produced high quality libraries regardless of sample quality or input [32]. Also accepted a similar level of mapping rates in his study with Q30 scores with >98% and >95% of the clean reads. On a per library basis, the proportions of the clean reads mapped to known Musa acuminata genome sequences databases (ASM31385V1) as in Table 2 were successfully determined. All the raw sequence data were deposited in the NCBI Sequence Read Archive database under nine accession number SAMN03793159; SAMN03793160; SAMN03793161; SAMN03793162; SAMN03793163; SAMN03793164; SAMN03793165; SAMN03793166; SAMN03793167. However, the percentage of reads mapped to the reference genome were reduced from 2>48>96 h infection.
| Sample ID | Total read bases | Total read pairs | Trimmed reads | Trimmed nucleotides | GC % | Q30 | Mapped Reads | Overall read mapping % | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Left-end | right end | |||||||||
| 2 h inoculation | Zero 1 D0 9889-6 DAY0-RNA1b | 7,056,367,222 4,871,369,736 6,859,348,744 | 69,865,022 50,813,776 67,914,344 | 5,968,001 2,176,010 3,961,035 | 1,526,127,066 857,017,236 1,068,208,967 | 49.47 51.63 51.00 | 89.16 93.94 91.05 | 34932511 15530826 11840237 | 34 932 511 15586502 11729203 | 54.60 62.80 34.87 |
| 48 h inoculation | D2-1 D2 9889 5 Day 2 9889 tube 2 | 7,740,380,834 4,690,276,424 4,606,319,928 | 76,637,434 45,607,128 45,607,128 | 5,597,388 2,039,911 3,616,370 | 1,391,673,770 787,842,199 860,384,321 | 52.15 50.97 51.06 | 90.34 94.14 89.83 | 1771549 15573245 3183370 | 1723470 15620946 3112431 | 4.60 65.5 13.80 |
| 96 h inoculation | D4-3 Day4 9889 G DAY 4 9889 tube 2 | 764,394,976 4,715,314,252 4,068,614,310 | 75,657,376 49,949,004 40,283,310 | 5,436,362 3,721,145 3,186,221 | 1,346,409,784 963,894,822 766,772,363 | 51.35 51.83 51.12 | 90.49 93.48 89.72 | 55137 11427740 149869 | 53314 11451970 147035 | 0.10 47.60 0.70 |
| *Q20;Q30 means base quality more than 20 and 30 respectively | ||||||||||
Table 2: Output statistics of the raw transcriptome and reference mapping of Musa acuminata cv. ‘Berangan’ infected with FocR4.
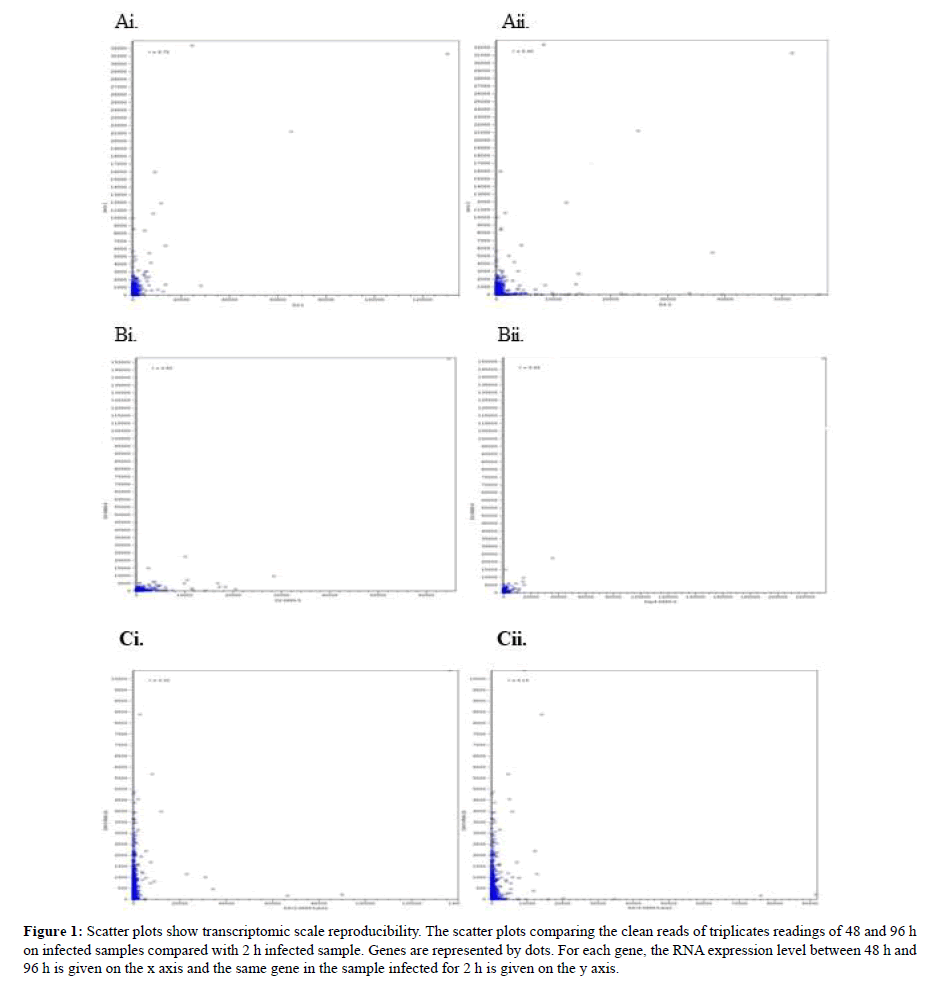
Scatter Plot Analysis
Scatter plots of these data are shown in Figure 1. These scatter plots showed that the experimental genes exhibit less variation overall. However, sample B showed less variation compared to sample A and C. By using the R value from the graph prediction on the accurate infection time which was predicted showed that the R value from graph Ai, Bi and Ci which derived from sample infected for 48 h showed high R value >0.5 than sample infected for 96 h which displayed low R value. This concludes that samples infected with shorter time length will produce significant R value than infection samples for lengthy time. The present findings seem to be consistent with other research which found the most genes exhibit less variation in expression between the biological duplicates when compared to the scatter plots between treatments [33]. Our findings are in agreement with [34] findings which showed that detection P-values (<0.05) showed lower reproducibility. To further confirm daya reproducibility, PCA plot was plotted.
Figure 1: Scatter plots show transcriptomic scale reproducibility. The scatter plots comparing the clean reads of triplicates readings of 48 and 96 h on infected samples compared with 2 h infected sample. Genes are represented by dots. For each gene, the RNA expression level between 48 h and 96 h is given on the x axis and the same gene in the sample infected for 2 h is given on the y axis.
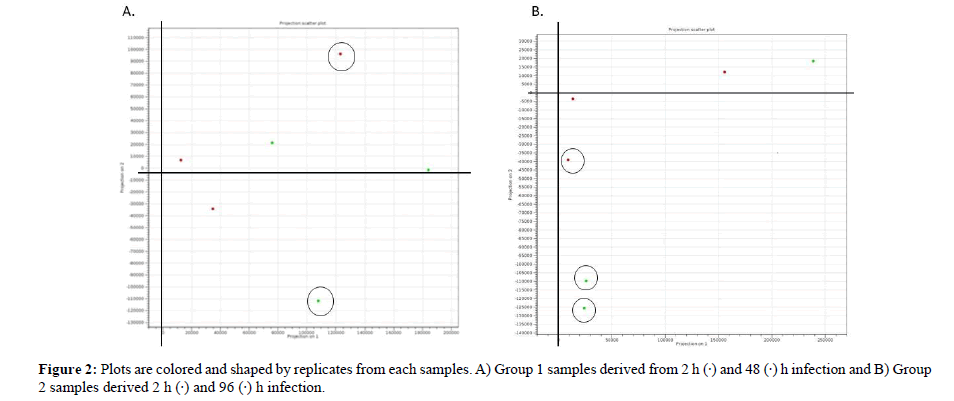
PCA Plot Analysis
PCA is a tool for identifying the main axes of variance within a data set and allows for easy data exploration to understand the key variables in the data and spot outliers. Properly applied, it is one of the most powerful tools in the data analysis tool kit [35]. PCA plot based on 3 biological replicates within each group resulted in a clear separation and large differences among group 1 between 2- and 48-h infection and group 2: Between 2- and 96 h infection. The PCA plot captures the variance in a dataset in terms of principal components and displays the most significant of data on the x, and y axes [35,36]. The percentages of the total variation are accounted for by the 1st and 2nd principal components which are shown on the x-, and y-axes labels [37]. From the PCA result we conclude that 96 h infection has produced 3 outliers than 48 h infection that produced 2 outliers therefore early detection is far better as it will provide more significant data for interpretation. In addition, the triplicate treatments of 96 h infection are more scattered than those of the sample infected for 48 h. A possible explanation for this might be that early diagnosis in plant infection are convincing.
Figure 2: Plots are colored and shaped by replicates from each samples. A) Group 1 samples derived from 2 h (·) and 48 (·) h infection and B) Group 2 samples derived 2 h (·) and 96 (·) h infection.
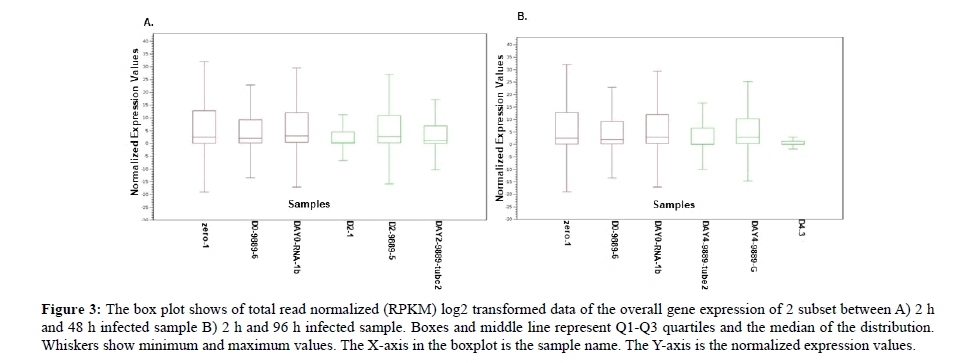
Box-Plot Analysis
Box plot normalized intensity values for each sample as shown in the box plot all samples carry average values lower than 15 and higher than 0 therefore all samples are included for further analysis (Figure 3). The median and the quartile values between the two groups were identical and most of the samples fell below the upper quartile. In addition, the median falls towards the lower quartile showing a positive skew (skewed right) and in sample D2_1, DAY 4_9889-Tube 2 and D4_3 the notches in the boxplots do overlap. This showed lack of 95% confidence and the true medians do not differ while on the other hand the remaining sample showed 95% confidence with true medians. Therefore, the expression levels of infected samples for 48 h and 96 h are varied suggesting that infection time length does play a major role in the regulation of the transcriptome. This result differs from who reported a symmetric pattern but is consistent with published data in who reported skewed right pattern [38,39]. These findings may have helped us to understand that plant defence varies in different exposures.
Figure 3: The box plot shows of total read normalized (RPKM) log2 transformed data of the overall gene expression of 2 subset between A) 2 h and 48 h infected sample B) 2 h and 96 h infected sample. Boxes and middle line represent Q1-Q3 quartiles and the median of the distribution. Whiskers show minimum and maximum values. The X-axis in the boxplot is the sample name. The Y-axis is the normalized expression values.
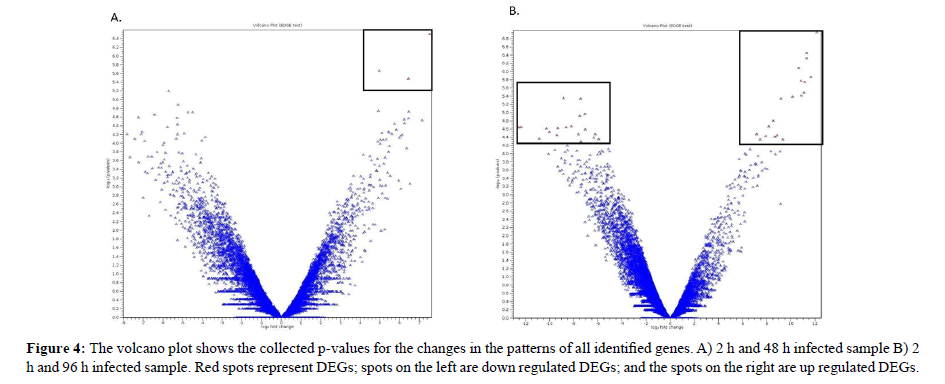
Volcano Plot Analysis
Volcano plot Figure 4a and 4b showed the differentially expressed genes. The volcano plot arranges genes along dimensions of biological and statistical significance [40]. The horizontal dimension corresponds to the biological impacts of the fold change between the two experimental groups on a log scale that meant for up and down regulation in a symmetric form [41]. While, the vertical axis represents the statistical evidence as p-value for a t-test of differences between experimental samples which is on a negative log scale and smaller p-values will appear higher up [42]. The dotted boxed showed significantly differential genes in infected samples for 48 and 96 h compared to 2 h of infection. These unigenes showed a significant up regulated and down regulated gene expression. The results clearly showed that fewer genes were detected in samples infected for 48 h and high numbers of genes were detected in samples infected for 96 h. Therefore, samples infected with less time will provide a smaller number of genes that were regulated which can be used as a marker to identify the infection at an early stage. These candidate marker genes groups were utilized for further analysis. Statistical analysis in the volcano plot analysis further reduced the number of differentially expressed genes based on a two combination comparison between 48 h and 96 h infection time point at p<0.05 and fold change 2.0 and above which display the significant transcript concentration [43].
Annotation and Classification of Predicted Proteins
To annotate and classify the 32709 unigenes of the infected samples, BLASTx, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were utilized. BLASTx was used to match them using the non-redundant (nr) protein database from NCBI, GO and KEGG database (cut-off e-value < 0.00001) [44]. From the results, 3 unigenes from sample inoculated for 48 h and 38 genes from sample infected for 96 h were successfully annotated through Swiss-Prot and functionally annotated. Through KEGG, one unigene and 9 unigenes were categorized under three major domains such as biological processes, cellular components and molecular function (Tables 3 and 4). The rest of the unigenes were reported as unidentified genes function. Only small numbers of unigenes were matched to known genes even though the e-value distribution of the top hits in the nr database showed that the mapped sequences display a certain level of homology. It is because due to some shortfalls that need to be considered such as only a small amount of RNA content was present from the infected plant roots and it appears as short sequences due to degradation through infection and limitation of the sequence information availability [45]. This problem can be overcome further validating through manual approaches or wet laboratory analysis simultaneously [46].
| Feature ID | Annotations - SwissProt | Annotations - Ontology | Annotations Pathway |
|---|---|---|---|
| LOC103969247 | ACT_GOSHI | ND | ND |
| LOC103969776 | DIR19_ARATH | GO:0048046 // cellular_component // apoplast | ND |
| LOC103974217 | FOMT2_WHEAT | ND | ND |
| *ND: Not detected | |||
Table 3: Unigenes detected from sample inoculated for 48 h.
| Feature ID | Annotations-SwissProt | Annotations - Ontology | Annotations - Pathway |
|---|---|---|---|
| LOC103969247 | ACT_GOSHI | . | . |
| LOC103969590 | ACT_GOSHI | . | . |
| LOC103970428 | MY108_ARATH | GO:0005634 // cellular_component // nucleus /// GO:0003682 // molecular_function // chromatin binding /// GO:0003677 // molecular_function // DNA binding /// GO:0003700 // molecular_function // sequence-specific DNA binding transcription factor activity /// GO:0006952 // biological_process // defense response /// GO:0009737 // biological_process // response to abscisic acid /// GO:0009723 // biological_process // response to ethylene /// GO:0009620 // biological_process // response to fungus /// GO:0009753 // biological_process // response to jasmonic acid /// GO:0009651 // biological_process // response to salt stress /// GO:0006351 // biological_process // transcription, DNA-templated | . |
| LOC103970628 | H4_SOYBN | GO:0009507 // cellular_component // chloroplast /// GO:0005829 // cellular_component // cytosol /// GO:0005730 // cellular_component // nucleolus /// GO:0000786 // cellular_component // nucleosome /// GO:0005886 // cellular_component // plasma membrane /// GO:0009506 // cellular_component // plasmodesma /// GO:0009579 // cellular_component // thylakoid /// GO:0005774 // cellular_component // vacuolar membrane /// GO:0003677 // molecular_function // DNA binding /// GO:0006334 // biological_process // nucleosome assembly | map05034 /// map05203 /// map05322 |
| LOC103971432 | R27AA_ORYSJ | . | map03010 |
| LOC103971565 | ERF71_ARATH | GO:0005634 // cellular_component // nucleus /// GO:0003677 // molecular_function // DNA binding /// GO:0003700 // molecular_function // sequence-specific DNA binding transcription factor activity /// GO:0009873 // biological_process // ethylene-activated signaling pathway /// GO:0034059 // biological_process // response to anoxia /// GO:0006351 // biological_process // transcription, DNA-templated | . |
| LOC103971966 | PME41_ARATH | . | map00040 /// map00500 /// map01100 |
| LOC103972067 | DOF46_ARATH | . | . |
| LOC103972251 | FLS_PETCR | . | . |
| LOC103972345 | PDC2_ORYSI | . | . |
| LOC103972882 | ACT_GOSHI | . | . |
| LOC103975493 | ACT_GOSHI | . | . |
| LOC103975908 | S47A1_PONAB | . | . |
| LOC103976080 | ERF26_ARATH | GO:0005634 // cellular_component // nucleus /// GO:0003677 // molecular_function // DNA binding /// GO:0003700 // molecular_function // sequence-specific DNA binding transcription factor activity /// GO:0009873 // biological_process // ethylene-activated signaling pathway /// GO:0006351 // biological_process // transcription, DNA-templated | . |
| LOC103976586 | . | . | . |
| LOC103977072 | ACT_GOSHI | . | . |
| LOC103977364 | . | . | . |
| LOC103979306 | ACT_GOSHI | . | . |
| LOC103979724 | H4_SOYBN | GO:0009507 // cellular_component // chloroplast /// GO:0005829 // cellular_component // cytosol /// GO:0005730 // cellular_component // nucleolus /// GO:0000786 // cellular_component // nucleosome /// GO:0005886 // cellular_component // plasma membrane /// GO:0009506 // cellular_component // plasmodesma /// GO:0009579 // cellular_component // thylakoid /// GO:0005774 // cellular_component // vacuolar membrane /// GO:0003677 // molecular_function // DNA binding /// GO:0006334 // biological_process // nucleosome assembly | map05034 /// map05203 /// map05322 |
| LOC103981239 | TBB1_LUPAL | . | . |
| LOC103982255 | TIF5A_ARATH | GO:0005634 // cellular_component // nucleus /// GO:0006952 // biological_process // defense response /// GO:0006355 // biological_process // regulation of transcription, DNA-templated /// GO:0006351 // biological_process // transcription, DNA-templated | . |
| LOC103982274 | ACT_GOSHI | . | . |
| LOC103983081 | . | . | . |
| LOC103983307 | TBB1_LUPAL | . | . |
| LOC103984614 | TBB7_GOSHI | . | . |
| LOC103984705 | ACCO1_ARATH | . | map00270 /// map01100 /// map01110 |
| LOC103986095 | ZAT12_ARATH | GO:0005634 // cellular_component // nucleus /// GO:0046872 // molecular_function // metal ion binding /// GO:0003700 // molecular_function // sequence-specific DNA binding transcription factor activity /// GO:0009631 // biological_process // cold acclimation /// GO:0042538 // biological_process // hyperosmotic salinity response /// GO:0009643 // biological_process // photosynthetic acclimation /// GO:0010200 // biological_process // response to chitin /// GO:0009409 // biological_process // response to cold /// GO:0009408 // biological_process // response to heat /// GO:0009416 // biological_process // response to light stimulus /// GO:0006979 // biological_process // response to oxidative stress /// GO:0010224 // biological_process // response to UV-B /// GO:0009611 // biological_process // response to wounding /// GO:0006351 // biological_process // transcription, DNA-templated | . |
| LOC103986585 | TBA_PRUDU | . | . |
| LOC103986777 | ACT_GOSHI | . | . |
| LOC103987214 | C94C1_ARATH | . | . |
| LOC103987224 | MKKA_DICDI | . | . |
| LOC103988107 | H32_WHEAT | GO:0000786 // cellular_component // nucleosome /// GO:0005634 // cellular_component // nucleus /// GO:0003677 // molecular_function // DNA binding | . |
| LOC103991389 | ACT_GOSHI | . | . |
| LOC103992122 | TBA_PRUDU | . | . |
| LOC103992213 | EF109_ARATH | GO:0005634 // cellular_component // nucleus /// GO:0003677 // molecular_function // DNA binding /// GO:0003700 // molecular_function // sequence-specific DNA binding transcription factor activity /// GO:0050832 // biological_process // defense response to fungus /// GO:0009873 // biological_process // ethylene-activated signaling pathway /// GO:0010200 // biological_process // response to chitin /// GO:0006351 // biological_process // transcription, DNA-templated | . |
| LOC103997416 | ACT_GOSHI | . | . |
| LOC103997697 | . | . | . |
| LOC103999973 | ERF20_ARATH | GO:0005634 // cellular_component // nucleus /// GO:0003677 // molecular_function // DNA binding /// GO:0003700 // molecular_function // sequence-specific DNA binding transcription factor activity /// GO:0009873 // biological_process // ethylene-activated signaling pathway /// GO:0010200 // biological_process // response to chitin /// GO:0006351 // biological_process // transcription, DNA-templated | . |
Table 4: Unigenes from sample inoculated for 96 h.
Pathway Analysis
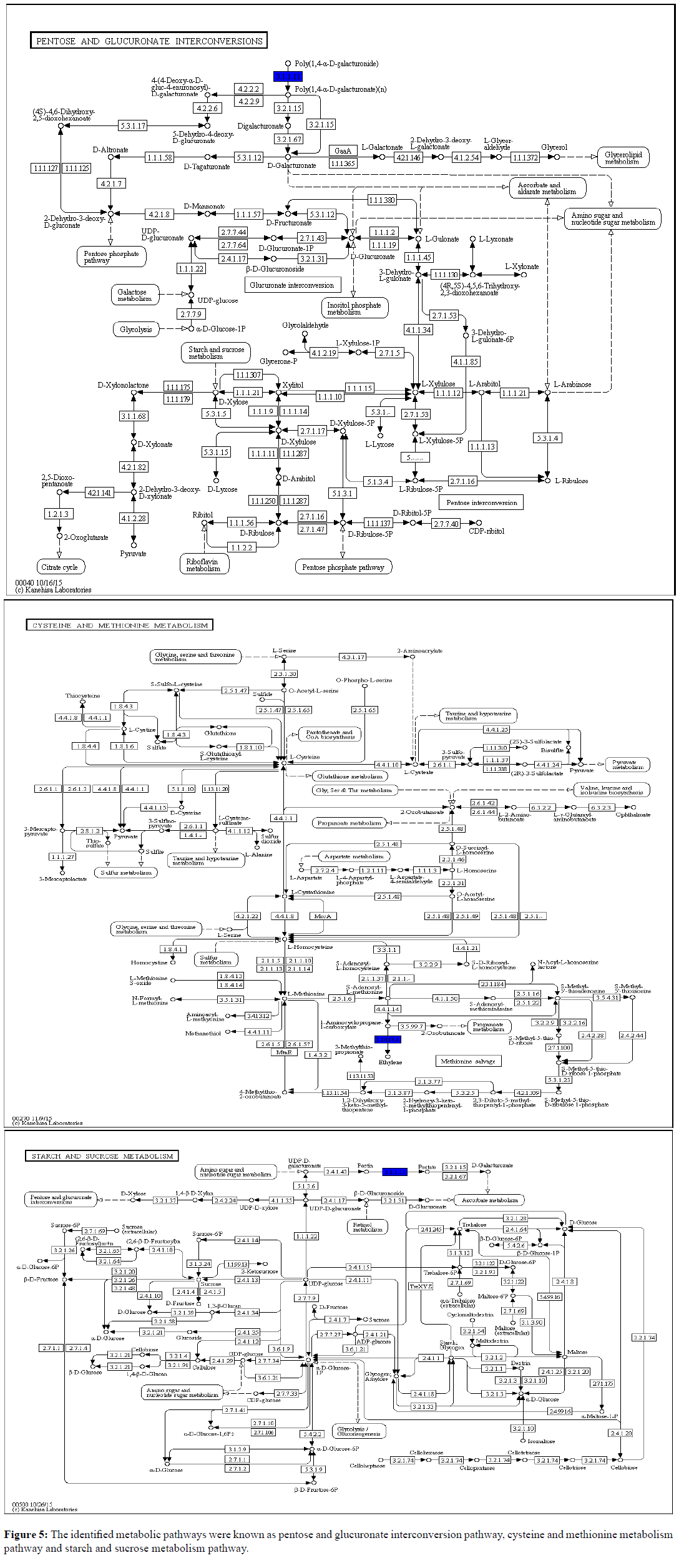
Pathway Analysis (PA), also known as functional enrichment analysis, is a fast and foremost tool in omics research. It interprets the differential expression results in terms of biological processes or molecular pathways. It uses the gene ontology resource databases to annotate genes based on an annotation dictionary [47]. The main purpose of PA tools is to analyze data obtained from high-throughput technologies then detecting the relevant groups of related genes that are altered in the experimental samples with comparison to a control [48]. In our results Figure 5, the identified metabolic pathways were known as Pentose and Glucuronate Interconversion Pathway, Cysteine and Methionine Metabolism Pathway and Starch and Sucrose Metabolism Pathway. Pentose and Glucuronate Interconversion and Starch Sucrose metabolism are the primary carbohydrate metabolism pathways. This shows that during an early infection most carbohydrate pathways were initiated to induce PR genes. In Arabidopsis, the induction of PR-1 and PR-5 by glucose was demonstrated in liquid cultures [49]. This explanation was further proved by current reported data when various PR genes were revealed consequently triggering carbohydrate metabolism for early defense. Together, these results suggest that carbohydrate metabolism positively will regulate the expression of defense-related genes. Moreover, cysteine which occupies a central position in plant primary and secondary metabolism together with methionine closely linked to hormone ethylene to be involved in modulation of plant responses to stresses [50,51]. Ethylene is synthesized in the cytosol from methionine via S-adenosyl-L-methionine (SAM), which is converted to 1-aminocyclopropane-1-carboxylic acid (ACC), and ACC is converted to ethylene and this further explained ACC identification in this study [52].
Real Time Analysis
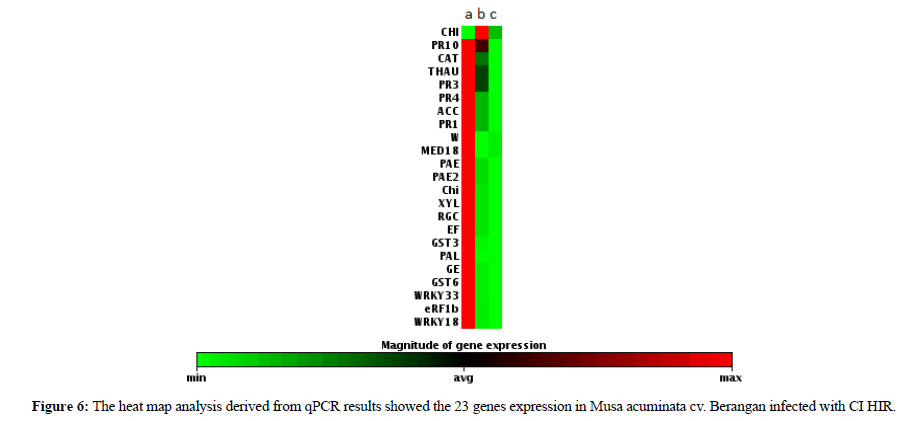
In this analysis all the data was handled independently and was normalized by using housekeeping gene Ribosomal Protein S2 (RPS2) [20]. The data was reported in terms of fold change which the expression was carried out through gene by gene by comparing the normalized Ct values (∆Ct) of all the biological replicates between three groups of samples [53]. The qPCR results of the 22 selected genes showed that all genes were expressed in 2 h of infection (Figure 6). On the other hand, the CHI gene started to be expressed in 48 h and was not detected in 96 h of infection. From this data we noticed that among the 22 genes that were tested only Chitinase was able to show a significant identification exclusively expressed in 48 h of infection. The result provided us with a new clue to understand that the early pathogenesis identification can be solved by using chitinase genes. Chitinase is the major fungal degrading enzyme produced by plants. Once attacked by a pathogen, the plant will release chitinase to degrade the fungal cell wall that mostly consists of chitin [54]. It is the first line defense that includes modification of the physical barriers such as cuticles and cell wall. Hence, chemical barriers such as phytoanticipins, saponins, phenols, quinines, defensins, peptides and proteins represent the second line defence [55]. This summarizes that CHI genes exhibit a unique characteristic than the rest of the tested gene. Therefore, CHI gene can be utilized for early diagnosis of fusarium infection.
| Genes | Primers | Tm °C | Nucleotide sequence 5` to 3` | Applications |
|---|---|---|---|---|
| Pectin acetylesterase-2 | PAE2F | 60 | GGCTCTCCTTTCTGGATGTTC | qPCR |
| PAE2R | 64 | TCAGCAAGGCACTTGACTTTT | qPCR | |
| Pectin acetylesterase | PAEF | 60 | GGCTCTCCTTTCTGGATGTTC | qPCR |
| PAER | 64 | TCAGCAAGGCACTTGACTTTT | qPCR | |
| Resistance Gene Candidates | RGC1F | 56 | CAAGTCTTGTCGAATCGAAC | qPCR |
| RGC1R | 60 | TCGTCGGCATGCCAGAATAC | qPCR | |
| WRKY transcription factors | WF | 53 | CCAGATACTTCGTGGATTGAAG | qPCR |
| WR | 53 | AGACATCAATAGCTGCAGTG | qPCR | |
| WRKY transcription factors | WRKY33F | 56 | GTGATATTGACATTCTTGACGA | qPCR |
| WRKY33R | 60 | GTGATATTGACATTCTTGACGA | qPCR | |
| WRKY transcription factors | WRKY18F | 57 | CGAAGGAGGAGGTCAAGGTT | qPCR |
| WRKY18R | 55 | TGGTGATGTAGTGCGTAGTAGT | qPCR | |
| Elongation Factor | EF-F | 57 | AACCCCCAAATATTCCAAGG | qPCR |
| EF-R | 61 | AGATTGGCACGAAAGGAATC | qPCR | |
| Chitinase | CHIF | 55 | CACCATCTCCTGCAAGCATA | qPCR |
| CHIR | 55 | GCAGTCATTCCTCGTTGTCA | qPCR | |
| Thaumatin-like protein | THAUF | 59 | CCGGTGGGCTAATTACAGG | qPCR |
| THAUR | 60 | CAATTCGGATGTCAATGCAG | qPCR | |
| Pathogenesis-related protein PR-3 | PR3F | 58 | GTCACCACCAACATCATCAA | qPCR |
| PR3R | 61 | CCAGCAAGTCGCAGTACCTC | qPCR | |
| Pathogenesis-related protein PR-3 | PR4F | 54 | CAGAACATTAACTGGGATTTGAGAG | qPCR |
| PR4R | 55 | CTCCATTTGCTGCATTGATCTACT | qPCR | |
| Pathogenesis-related protein PR-1 | PR1F | 57 | TCCGGCCTTATTTCACATTC | qPCR |
| PR1R | 61 | GCCATCTTCATCATCTGCAA | qPCR | |
| Pathogenesis-related protein PR-10 | PR10F | 60 | CTCCGAGAAGCAGTACTACGA | qPCR |
| PR10R | 62 | GATGGCCGTGGACGAA | qPCR | |
| Phenylalanine ammonia lyase | PALF | 63 | ACAGGAGGACCAAGCAAGGA | qPCR |
| PALR | 64 | CGTCCCGGAGCCGAATAT | qPCR | |
| Catalase | CATF | 63 | AAGGTCTCACCGCTTGTCTCA | qPCR |
| CATR | 64 | CGTCGCGGATGAAGAACAC | qPCR | |
| 40s Ribosomal Protein | RPS2F | 60 | TAGGGATTCCGACGATTTGTTT | qPCR |
| RPS2R | 63 | TAGCGTCATCATTGGCTGGGA | qPCR | |
| Aminocyclopropae carboxylic acid | ACCF | 54 | AAGATGGCACTAGGATGTCAATAG | qPCR |
| ACCR | 54 | TCCTCTTCTGTCTTCTCAATCAAC | qPCR | |
| Mediator18 | MED18F | 55 | TTCCTGTAACACCTGGTATGC | qPCR |
| MED18R | 55 | GGAGATAGACGGTTTCGACAAG | qPCR | |
| Chitinase | ChiF | 60 | CCCAATTTCTTTCGCCGCTATGCT | qPCR |
| ChiR | 60 | TGTTCGGCTCTCATGACCTTCTCA | qPCR | |
| Xylanase | XYLF | 62 | GCGCCGGCGGTGAT | qPCR |
| XYLR | 55 | GATAAACCCGAGCCGCTTCT | qPCR | |
| Glutathione S-transferases | GST3F | 55 | ATGGCTTGGGTCAAGAGATG | qPCR |
| GST3R | 53 | CCAACCCACACAACCATAG | qPCR | |
| Germin Family Protein | GEF | 49 | TTCCTCTTTGCTCTTGTC | qPCR |
| GER | 50 | AGTGTTTGTGGTGTTTCC | qPCR | |
| Glutathione S-transferases | GST6F | 55 | TCATCAACCACCCTGTTGTC | qPCR |
| GST6R | 51 | AAATGGAAACAAGATCCAAGG | qPCR | |
| Eukaryotic release factor | eRF1bF | 59 | TCATTCTCTTGAAGTTGGGGCATTAGATCT | qPCR |
| eRF1bR | 55 | CTCGTTCTTGAAGTATTTTGAATCTTTTTCC | qPCR |
Table 5: List of primers.
Conclusion
The signaling pathways generated in the present study revealed that the defense system of bananas is complex and in depth understanding of the banana defense response to plant pathogens are crucial. Many defense related genes and pathways in bananas differ from model plants suggesting that the mechanism underlying host defense in plants are variable. Among the generated sequences, unigenes that were specifically expressed could play an important role in the interaction of banana and fusarium. It will provide insight into the evolution of the pathogenic processes. Our study provides a substantial contribution to the existing number of the deposited data and resources to the success in combating banana infection. The findings of this study will accelerate research on finding tolerance banana varieties towards fusarium.
Declarations
Acknowledgements
This work was supported by grant (53-02-03-1068) financed by the Ministry of Science, Technology and Innovation (MOSTI) Malaysia, a University of Malaya Research Grant (RP005C-13BIO) and a University Malaya High Impact Research Grant (UM.C/625/1/HIR/MOHE/SCI/18). This work was edited by Lim Yee Meng and Wee Ben Jie.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Swarupa V, Ravishankar KV, Rekha A. Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta. 2014, 239:735-751.
- Tan EES, Abdullah A, Musa KH, Mohammad YM, Ghani MA. Antioxidant properties of three banana cultivars (Musa acuminata ‘Berangan’, ‘Mas’ and ‘Raja’) extracts. Sains Malaysiana, 2012, 41:319-324.
- Ding D, Zhang L, Wang H, Liu Z, Zhang Z, et al. Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot, 2009, 103:29-38.
- Mohd Rasidek NA, Mad Nordin MF, Shameli K. Formulation and evaluation of semisolid jelly produced by Musa acuminata Colla (AAA Group) peels. Asian Pacific J Trop Biomed. 2015, 6:155-159.
- Heslop-Harrison JS, Schwarzacher T. Domestication, genomics and the future for banana. Ann Bot. 2007, 100:1073-1084.
- Subramaniam S, Maziah M, Sariah M, Puad MP, Xavier R. Bioassay method for testing Fusarium wilt disease tolerance in transgenic banana. Scientia Horticulturae, 2006, 108:378-389.
- Thangavelu R, Perumal GD, Mustaffa M, Sreeramanan S, Rathinam X. Genomics of Fusarium oxysporum f. sp. cubense causing wilt disease in Banana (Musa spp.) In: Genetics, Genomics and Breeding of Bananas. Taylor & Francis Group. 2012, pp:1-324.
- Butler D. Fungus threatens top banana. Nature. 2014, 7479:195-196.
- IPPC-International Plant Protection Convention. New banana disease found in Mozambique (Fusarium oxysporum f.sp. cubense tropical race 4). Retrieved June 2, 2014.
- García-Bastidas F, Konkol J, Al-Qasim M, Naser Z, Abdelwali M, et al. First report of Fusarium oxysporum f. sp. cubense tropical race 4 associated with panama disease of banana outside Southeast Asia. Disease Notes. 2014, 98:5-694.
- Ortiz R, Swennen R. From crossbreeding to biotechnology-facilitated improvement of banana and plantain. Biotechnol Adv. 2014, 32:158-169.
- Biosecurity Queensland. Panama disease tropical race 4: Standards and guidelines. 2015.
- Li WM, Dita M, Wu W, Hu GB, Xie JH, et al. Resistance sources to Fusarium oxysporum f. sp cubense tropical race 4 in banana wild relatives. Plant Pathol. 2014.
- Huang J, Gu M, Lai Z, Fan B, Shi K, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development and response to environmental stress. Plant Physiol. 2010, 153:1526-1538.
- D'Hont AF, Denoeud JM, Aury FC, Baurens F, Carreel O et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012, 488:213-217.
- Davey ML, Heimdal R, Ohlson M, Kauserud H. Host and tissue-specificity of moss-associated Galerina and Mycena determined from amplicon pyrosequencing data. Fungal Eco. 2013, 6:179-186.
- Passos MAN, De Cruz VO, Emediato FL, De Teixeira CC, Azevedo VCR, et al. Analysis of the leaf transcriptome of Musa acuminata during interaction with Mycocphaerella musicola: Gene assembly, annotation and marker development. BMC Genomics. 2013,14:78-95.
- Li X, Bai T, Li Y, Ruan X, Li H. Proteomic analysis of Fusarium oxysporum f. sp. cubense tropical race 4-inoculated response to Fusarium wilts in the banana root cells. Proteome Sci. 2013,11.
- Thakker JN, Dalwadi P, Dhandhukia PC. Biosynthesis of gold nanoparticles using Fusarium oxysporum f. sp. Cubense JT1, a plant pathogenic fungus. ISRN Biotechnol. 2013, 515091:5.
- Munusamy U, Mohd-Yusuf Y, Baharum NA, Zaidi K, Othman RY. RT-qPCR profiling of pathogenesis related genes in Musa acuminata cv. Berangan seedlings challenged with Fusarium oxysporum f. sp. cubense tropical race 4. Pak J Agri Sci. 2019, 56:37-42.
- Baharum NA, Othman RY, Mohd-Yusuf Y, Tan BC, Zaidi K, et al. The effect of pathogenesis-related 10 (Pr-10) gene on the progression of Fusarium wilt in Musa acuminata cv. Berangan (kesan gen berkaitan patogenesis 10 (Pr-10) pada perkembangan Fusarium layu dalam Musa acuminata cv. Berangan). Sains Malaysiana. 2018, 47:2291-2300.
- Ting ASY, Sariah M, Kadir J, Gurmit S. Field evaluation of non-pathogenic Fusarium oxysporum isolates UPM31P1 and UPM39B3 for the control of fusarium wilt in 'pisang berangan' (Musa, AAA). Acta Hortic. 2009, 828:139-144.
- Gubbuk H, Erkan M, Onus AN. Identification and selection of superior banana phenotypes in the cultivar Dwarf Cavendish using agronomic characteristics and RAPD markers. Pak J Bot. 2004, 36:331-342.
- Ma Y, Liu M, Stiller J, Liu C. A pan-transcriptome analysis shows that disease resistance genes have undergone more selection pressure during barley domestication. BMC Genomics. 2019, 20:12.
- Guo L, Han L, Yang L, Zeng H, Fan D, et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. cubense causing banana vascular wilt disease. PLoS ONE. 2014, 9(4):e95543.
- Kimbrel JA, Di Y, Cumbie JS, Chang JH. RNA-Seq for Plant Pathogenic Bacteria. Genes (Basel). 2011, 2:689-705.
- Dang Y, Xu X, Shen Y, Hu M, Zhang M, et al. Transcriptome analysis of the innate immunity-related complement system in spleen issue of Ctenopharyngodon idella infected with Aeromonas hydrophila. PLoS ONE. 2016, 11:e0157413.
- Munusamy U, Mohd-Yusuf Y, Baharum NA, Zaidi K, Othman RY. Evaluation of pathogenicity level of three different strains of Fusarium oxysporum in Musa acuminata cv. Berangan. Res J Biotechnol. 2019, 14:12-20.
- Zhou J, Wang M, Sun Y, Gu Z, Wang R, et al. Nitrate increased cucumber tolerance to Fusarium wilt by regulating fungal toxin production and distribution. Toxins (Basel). 2017, 9:100.
- Quail MA, Swerdlow H, Turner DJ. Improved protocols for the Illumina genome analyzer sequencing system. Curr Protoc Hum Genet. 2009, Chapter 18: Unit 18.2.
- Ledergerber C, Dessimoz C. Base-calling for next-generation sequencing platforms. Brief Bioinform. 2011, 12:489-497.
- Xiong H, Guo H. Xie Y, Zhao L, Gu J, et al. RNAseq analysis reveals pathways and candidate genes associated with salinity tolerance in a spaceflight-induced wheat mutant. Scientific Reports. 2017, 7:2731.
- Weeda S, Zhang N, Zhao X, Ndip G, Guo Y, et al. Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS One, 2014, 9:e93462.
- Kakei Y, Shimada Y. AtCAST3.0 Update: A web-based tool for analysis of transcriptome data by searching similarities in gene expression profiles. Plant Cell Physiol. 2015, 56:e7.
- Altman N, Krzywinski M. Nat Methods, 2017, 14:545-546.
- Todorov H, Fournier D, Gerber S. Review principal components analysis: Theory and application to gene expression data analysis. Genomics Computational Biol. 2018, 4:e100041.
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-Wide Analysis of a group Streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathogen. 2006, 2:e5.
- Garcia-Sanchez S, Bernales I, Cristobal S. Early response to nanoparticles in the Arabidopsis transcriptome compromises plant defence and root-hair development through salicylic acid signalling. BMC Genomics. 2015, 16:341.
- Yang L, Jin Y, Huang W, Sun Q, Liu F, et al. Full-length transcriptome sequences of ephemeral plant Arabidopsis pumila provides insight into gene expression dynamics during continuous salt stress. BMC Genomics. 2018, 19:717.
- Liang LR, Lu S, Wang X, Lu Y, Mandal V, et al. FM-test: A fuzzy-set-theory-based approach to differential gene expression data analysis. BMC Bioinformatics. 2006, 12:7:S7.
- Love MI, Huber W, Anders A. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15:550.
- Wentian Li. Volcano plots in analyzing differential expressions with mRNA microarrays. J. Bioinformatics Computational Biol. 2012, 10:1231003.
- Ghaffar MBA, Norliza AB, Pritchard J, Ford-Lloyd BV. Identification of candidate genes involved in brown planthopper resistance in rice using microarray analysis. J Trop Agric Fd Sc. 2016, 44:49-62.
- Jung WY, Lee SS, Kim CW, Kim HS, Min SR, et al. RNA-seq analysis and de novo transcriptome assembly of Jerusalem artichoke (Helianthus tuberosus Linne). PloS ONE. 2014, 9:e111982.
- Cardoso-Silva CB, Costa EA, Mancini MC, Balsalobre TWA, Canesin LEC, et al. De Novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS ONE. 2018, 9:e88462.
- Bolger ME, Arsova B, Usadel B. Plant genome and transcriptome annotations: From misconceptions to simple solutions. Brief Bioinform. 2018, 19:437-449.
- Hill DP, Smith B, McAndrews-Hill MS, Blake JA. Gene ontology annotations: What they mean and where they come from. BMC Bioinformatics. 2008.
- García-Campos MA, Espinal-Enríquez J, Hernández-Lemus E. Pathway analysis: State of the art. Frontiers Physiol. 2015, 6:383.
- Xiao W, Sheen J, Jang JC. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol. 2000, 44:451-461.
- Gotor C, Alvarez C, Bermúdez MA, Moreno I, García I, et al. Low abundance does not mean less importance in cysteine metabolism. Plant Signaling Behavior. 2010, 5:1028-1030.
- Iqbal N, Masood A, Khan MI, Asgher M, Fatma M, et al. Cross-talk between sulfur assimilation and ethylene signaling in plants. Plant Signal Behav. 2012, 8:e22478.
- Bleecker AB, Kende H. Ethylene: A gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000, 16:1-18.
- Goni R, García P, Foissac S. The qPCR data statistical analysis. Integromics White Paper. 2009, pp:1-9.
- Kumar M, Brar A, Yadav M, Chawade A, Vivekanand V, et al. Chitinases-potential candidates for enhanced plant resistance towards fungal pathogens. Agri. 2018, 8:88.
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr OpinPlant Biol. 2002, 5:325-331.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences