Contamination of River Nun at Amassoma, Bayelsa State, Nigeria Due to Microbial Diversity in Sediments
Lovet T Kigigha, Vivian J Obua and Sylvester C Izah*
Department of Biological Sciences, Niger Delta University, Wilberforce Island, Bayelsa State, Nigeria
- *Corresponding Author:
- Sylvester C Izah
Department of Biological Sciences
Faculty of Science
Niger Delta University
Wilberforce Island, Bayelsa State, Nigeria
Tel: +234 7030192466
E-mail: chivestizah@gmail.com
Received Date: January 06, 2017; Accepted Date: January 23, 2017; Published Date: February 04, 2018
Citation: Kigigha LT, Seiyaboh EI, Obua VJ, Izah SC (2018) Contamination of River Nun at Amassoma, Bayelsa State, Nigeria Due to Microbial Diversity in Sediments. Environ Toxicol Stud J 2:2.
Abstract
Environmental contamination is a major issue challenging its sustainability. This study evaluated the microbial contamination level in sediment from River Nun at Amassoma axises in Bayelsa state, Nigeria. Triplicate sediment samples were collected from 5 locations along River Nun at Amassoma axises in Bayelsa state, Nigeria. The samples were analyzed using standard microbiological techniques. Results showed that microbial density ranged from 1.80 – 7.33 x 106 cfu/g, 1.81 – 13.53 x 104 cfu/g and 1.56 – 8.67 x 105 cfu/g for total heterotrophic bacteria, total coliform and total fungi respectively. There was significant difference (P<0.05) among the various sampling stations. The tentative microbial isolates from the sediment were Staphylococcus aureus, E. coli, Pseudomonas, Streptococcus, Enterobacter, Corynebacterium, Salmonella, Bacillus, Micrococcus, Proteus, Aspergillus, Penicillium, Saccharomyces, Mucor, Rhizopus, Fusarium and Geotrichum species. The similarity of the microbial diversity between the various locations based on Sorenson qualitative index ranged from 60.87 – 85.71%, being above critical level of significance of 50% for similarity. The occurrence of this array of microbes at further suggests that the water is contaminated and as such it requires treatment prior to use.
Keywords
Microorganisms; Sediment; Water pollutant
Introduction
Sediments arise from the sedimentation of substances found in surface water. The materials are mainly from weathering processes, runoff after precipitation, direct waste/and other material deposition in the water bodies due to human action, wind etc. Sediment is found in different surface water resources including creek, creeklets, ponds, lakes, rivers, stream, etc. Sediments is made up of organic (viz: dead and decomposed organisms) and inorganic materials (including dust, eroded soil etc.) [1-3]. According to Seiyaboh et al. the inorganic component of the sediment exists in the form of fine material that has a diameter of<2 mm, and coarse substances that have diameter>2 mm which are mainly gravel and bedrocks. The authors further reported that organic components are the remains and decomposed animal matter, aquatic plants, leaves, flowers, pollen, and even whole branches and trees.
Sediment typically exists in two perspectives viz: suspended and deposited sediment [3]. According to Adesuyi et al. [4] and Seiyaboh et al. sediment is a major reservoir to surface water pollutants, hence contaminating water column and aquatic organisms. Since sediment is associated to water, its quality could be affected by physical, chemical and biological (including microbiological) characteristics of the surface water itself. According to Seiyaboh et al. sediment has relationship with water and as such, the ability of lives to thrive in water depends on the water and sediment quality.
The sediment quality tends to be affected by the drainage nature of an area which could be either natural or man-made [2]. The authors further reported that the drainage play essential role on water quality especially in the Niger Delta region of Nigeria. Typically, the Niger Delta region of Nigeria has seven major drainage systems which lie squarely in the wet equatorial climatic belt [2]. In Bayelsa state, several surface water resources which are called several names including creek, creeklets, pond, stream and rivers exists. Most of these surface water bodies have their origin from Nun River-a major tributary of River Niger.
Microbes found in environmental components such as surface water and soil could also be found in sediment. Specifically Agbabiaka and Oyeyiola reported that soil and sediments vary but they both provide varying environments for microbes to thrive, and they also play different roles in the wetland ecosystem. The authors further reported that sediment could be autochthonous or allochthonous, and that majority of the microorganisms found in soil are also present in sediment probably due to the effects of soil erosion. Furthermore, environmental factors tend to provide a suitable condition for microbial proliferation. Conditions such as pH, moisture, organic matter, aeration, temperature, nutrients etc. affect the distribution, abundance, and growth etc. of microbesin an environment.
Since, water quality is related to sediment quality, therefore both water and sediment play essential role to several life forms. The presence of pollutant in sediment could affect the aquatic organisms. According to Agbabiaka and Oyeyiola the enrichment of sediment is also brought about by deposition of organic matter which may be derived from many sources such as sewage, either discharged as domestic or industrial effluent, dumped into and/ or close to surface water. Ogbonna further reported that this may enhance microbial density in the recipient surface water due to increased nutrient availability [5].
In Bayelsa state, several studies have been carried on water quality including River Nun [6-9], Ikoli creek [10-12], Kolo creek [13-15], Epie creek [16-18], Sagbama creek [19], but most of these studies focused on water quality. Though, few other studies focused on the sediment quality such as on Epie creek (Seiyaboh et al.), Sagmaba creek (Seiyaboh et al.), Kolo creek [20]. Of these studies, information on the microbial quality is scanty. This study aimed at assessing the microbial quality of river Nun at Amassoma axises in the Niger Delta, Nigeria.
Materials and Methods
Study area
Amassoma is one of the communities in Southern Ijaw Local Government Area of Bayelsa state. Amassoma is the host community of Niger Delta University, and it’s approximately 30 km from Yenagoa-the Bayelsa state capital. Several economic activities that are carried out in the area include farming, fishing and logging. Wastes generated from anthropogenic sources are discharged into the water body. In addition, pier toilet system is built on the water ways [9,6]. The State lies in the sedimentary basin. During the end of wet season (viz: September/October), the water level rise and almost entering the buildings of the residents aligning the water bank. The climatic environment of the area is peculiar to other Niger Delta regions which have been widely described by authors [2,7,8,10,12,14,15,21-25].
Sampling techniques
Triplicate sediment samples were collected at 5 locations at Amassoma stretching from Ending Pele to College of Health Science waterside. The samples were packaged in sterile bottles, labeled and transported to the laboratory for microbiological analysis.
Enumeration of total heterotrophic bacteria counts
MacConkey agar, Nutrient Agar and Potatoes Dextrose Agar were used to enumerate total coliform, total heterotrophic bacteria count and total fungi. The media were prepared based on the manufacturers’ instruction. Enumeration of the microbial density was carried out following pour plate method previously described by Pepper and Gerba (2005) and Benson (2002) [26,27]. Serial dilution was carried out up to 10-6 and approximately 1.0 ml of the diluents was aseptically plated in sterile petri dishes. Later, the prepared media was poured accordingly. The agar plates were swirled (clock wise and anti-clockwise), and allowed to solidify and then incubated inverted at 37°C for 24- 48 (bacteria) 3-5 days (fungi). The resultant colonies were counted and expressed as colony forming units per gram of the sediment sample.
Tentative identification of the microbial isolates
Bacteria: The resultant colonies were streaked onto differential media and then subjected to biochemical test using the guide of Cheesbrough [28]. Furthermore, the samples were also streaked onto Blood Agar, Mannitol salt Agar, Salmonella-Shigella Agar, Levine’s eosin Methylene Blue (EMB). The distinct isolates from the various media were also streaked onto nutrient agar and further subjected to other biochemical test. The presence of yellowish pigments in Mannitol Salt Agar indicates Staphylococcus aureus and this was confirmed by streaking the colonies in nutrient agar and subjected to coagulase and gram reaction test. From the EMB agar, the presence of small nucleated colonies with greenish metallic sheen indicates E. coli, while large nucleated colonies without the metallic sheen is an indication of Enterbacter species [27]. From the blood agar, the presence of swarming and haemolytic characteristics after incubation depicts Proteus species and Streptococcus species respectively. Furthermore, the presence of black colonies in Salmonella-Shigella Agar after incubation indicates Salmonella sp. The positive isolates on Salmonella agar were prepared into slants in a Triple Sugar Iron Agar. The presence of cracks and blackening in the tube depicts Salmonella species [29]. The resultant colonies from the biochemical tests were compared with that of known taxa using Bergey’s Manual of Determinative Bacteriology scheme by Holt et al. [30].
Fungi: Macroscopic/colonial and microscopic characteristics were used for fungi identification. The scheme of Ellis et al. and Benson was used for the macroscopic identification [31]. While the wet mount preparation using Lactophenol cotton blue stain as indicator previously described by Pepper and Gerba; Benson was used for the microscopic identification.
Cultural, morphological, and physiological/biochemical characteristics (using carbon fermentation and assimilation techniques, glucose-peptone-yeast extract broth) previously Kurtzman and Fell [32] and have been applied by Iwuagwu and Ugwuanyi, Okoduwa et al., Izah et al. was used for yeast identification [33-37]. The resultant characteristics were compared with the scheme provided by Ellis et al. and Benson.
Statistical analysis
Statistical Package for Social Sciences software version 20 was used for the statistical analysis. A univariate analysis of variance was carried out and significance variation was determined at P=0.05. Where variation exists, post hoc-Tukey Honestly Significance Difference was carried out to show the source of the variation. Sorenson qualitative index by Ogbeibu was used to determine the similarity among the microbial diversity between the various locations at critical level of significance=50% [38].
Results and Discussion
Table 1 represents the microbial density of sediment from River Nun at Amassoma axises, Bayelsa state, Nigeria. The total heterotrophic bacteria counts ranged from 1.80 – 7.33 x 106 cfu/g. There was significant variation (P<0.05) among the various locations the samples were obtained. However, there was no significant difference (P>0.05) for mean of: locations A and D, and Locations B, C and E. Total coliform counts ranged from 1.81 – 13.53 x 104 cfu/g, being significantly different (P<0.05) among the various locations. Furthermore, means of location A, B and C were not significantly different (P>0.05). Total fungi counts ranged from 1.56 – 8.67 x 105 cfu/g. Basically there was significant difference (P<0.05) among the various locations. Means of the various locations for total fungi were not significantly different (P>0.05) for Locations A and B, and Locations C, D and E.
| Locations | total heterotrophic bacteria x 106 cfu/g | Total coliform x 104 cfu/g | Total Fungi x 105 cfu/g |
|---|---|---|---|
| A | 1.80 ± 0.07a | 2.45 ± 0.26a | 8.67 ± 2.93c |
| B | 5.93 ± 0.61b | 1.81 ± 0.14a | 8.07 ± 0.78bc |
| C | 7.10 ± 1.03b | 2.51 ± 0.19a | 1.78 ± 0.17ab |
| D | 1.86 ± 0.11a | 13.53 ± 1.24c | 1.56 ± 0.08a |
| E | 7.33 ± 0.90b | 8.63 ± 0.49b | 1.74 ± 0.18ab |
Data is expressed as mean ± Standard Error; Different alphabets (a, b, c) along the column indicate significant variation (P<0.05) according to Tukey Honestly Significance Difference
Table 1: Microbial density of sediment from River Nun at Amassoma axises, Bayelsa state, Nigeria.
Table 2 represents the diversity of microbial isolates found in the sediment of River Nun at Amassoma axises, Bayelsa state, Nigeria. The tentative microbial isolates identified include Staphylococcus aureus, E. coli, Pseudomonas, Streptococcus, Enterobacter, Corynebacterium, Salmonella, Bacillus, Micrococcus and Proteus species (bacteria), Aspergillus, Penicillium, Saccharomyces, Mucor, Rhizopus, Fusarium and Geotrichum species (fungi) while Streptococcus and Corynebacterium species occured only in locations B and C respectively. In addition, Salmonella species also occurred in only Location C. Geotrichum species were only present in locations B and C. While Saccharomyces species were found in Locations A and C. Other microbial diversity occurred in ≥ 3 locations.
| Microbes | A | B | C | D | E |
|---|---|---|---|---|---|
| Pseudomonas sp. | + | + | + | - | + |
| Enterobacter sp. | + | + | + | + | + |
| Bacillus sp. | + | - | + | - | + |
| Streptococcus sp. | - | + | - | - | - |
| Staphylococcus aureus | + | + | + | + | + |
| Corynebacterium sp. | - | - | + | - | - |
| Micrococcus sp. | + | - | - | + | + |
| E. coli | + | + | + | + | + |
| Proteus species | + | - | + | + | - |
| Salmonella sp. | - | - | + | - | - |
| Aspergillus sp. | + | + | + | + | + |
| Penicillium sp. | + | - | + | + | - |
| Saccharomyces sp. | + | - | + | - | - |
| Mucor sp. | - | + | + | + | - |
| Rhizopus sp. | + | + | - | - | + |
| Fusarium sp. | + | + | + | + | + |
| Geotrichum sp. | - | + | + | - | - |
Some of the isolates were only found in one sample among the triplicate samples from each location
Table 2: Tentative microbial isolates from sediment of River Nun at Amassoma axises.
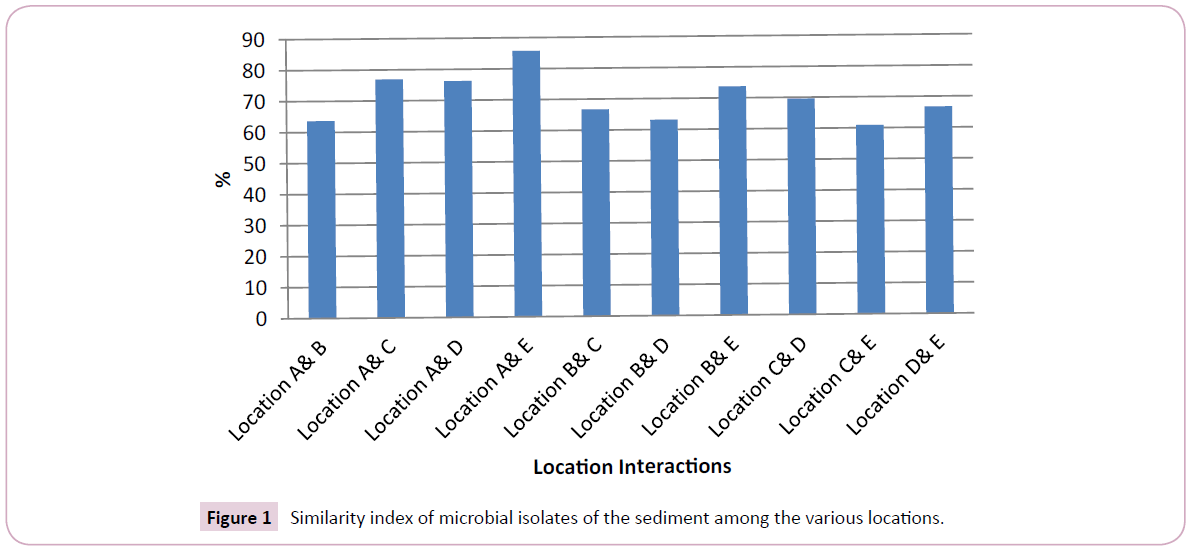
The similarity of the microbial diversity between the various location based on Sorenson qualitative index is presented in Figure 1. Based on microbial diversity, the similarity interaction between the various locations ranged from 60.87 – 85.71%, being above critical level of significance=50% for similarity.
The variation among the microbial density with regard to total heterotrophic bacteria, total coliform and total fungi counts could be associated to difference in the anthropogenic activities in the water. Seiyaboh et al. also reported difference in microbial density of the river at Amassoma axises which was attributed to variation in human activities prior to sampling [9]. The occurrences of coliforms could be due to the fact that, the surface water at Amassoma axises is a major recipient of different wastes up stream. Authors have reported that pier toilet system is built on water surfaces in most coastal communities in Bayelsa state [39,40] including Amassoma [7-9]. Authors have also reported that surface water in coastal region of Bayelsa state is a major receipt of wastes resulting from market activities [11,17,18] and runoff resulting from rainfall [16]. Furthermore, the high microbial counts could be associated to the presence of suspended or particulate solid rich in organic matter which may serve as nutrient sources [41].
The microbial density in this study had some similarity with the work of Ogbonna (2014) on sediment of surface water receiving abattoir wastes in Egbu in Imo State and Trans-Amadi in Port Harcourt, Rivers State [42]. Furthermore, Tijani et al. reported mean total heterotrophic bacteria and E. coli counts of 3.2 x 105 cfu/g and 6.0 x 103 cfu/g in sediment of Awba Lake, Ibadan, Oyo state, Nigeria [41]. But higher than the findings of Agbabiaka and Oyeyiola that reported density of microbial contaminant of sediment of Foma River, Ita-Nmo, Ilorin in the range of 4.0 x 102 to 2.03 x 104 cfu/g and 2.0 x 102 to 6.6 x 103 cfu/g for total bacteria and fungi counts, respectively [1].
Furthermore, the microbial density in the sediment of this study had some similarity with previous works on River Nun. Seiyaboh et al. reported to heterotrophic bacteria density in water samples from River Nun at Amassoma axises in the range of 1.78 to 9.30 x 106 cfu/ml [9]. Agedah et al. reported total heterotrophic bacteria in the range of 6.389 – 6.434 Log cfu/ml in some surface water around Wilberforce Island, Bayels state [7].
The microbial diversity reported in this study had some similarity with the values previously reported by some authors from sediment. For instance, Ogbonna have reported the presence of Klebsiella, Enterobacter, Acinetobacter, Citrobacter, Shigella, Achromobacter, Bacillus, Escherichia, Salmonella, Favobacterium, Lactobacillus, Micrococcus, Pseudomonas, Streptococcus, Serratia, Vibrio and Proteus species in sediment, soil and surface water receiving abattoir wastes [5]. Typically, abattoir wastes arises from condemned meat, undigested ingesta, bones, horns, hairs and aborted fetuses (solid wastes) and blood, gut contents, urine and water (liquid i.e. effluents) that are characterized by high suspended solid and dissolved solid containing liquid and fat [3]. Furthermore, Agbabiaka and Oyeyiola reported that microbes of the genera Corynebacterium, Escherichia, Bacillus, Enterobacter, Klebsiella, Staphylococcus, Micrococcus, Acinetobacter, Pseudomonas, Aeromonas, Salmonella, Streptococcus, Proteus and Erwinia (bacteria), Curvularia, Aspergillus, Penicillium, Saccharomyces, Cladosporium, Geotrichum, Trichoderma, Mucor, Rhizopus, Fusarium and Mortierella (fungi) as sediment contaminants of Foma River, Ita-Nmo, Ilorin [1].
The diversity of bacteria reported in this study has been reported in the surface water of river Nun by Seiyaboh et al. [9]. Furthermore, the various species reported in this study have been reported in Epie creek water receiving several wastes streams resulting from human activities as reported by Ben-Eledo et al. [17]. Also, the bacteria species in this study have been recorded (Staphylococcus aureus, E. coli, Pseudomonas, Enterobacter, Yersia, Shigella, Bacillus, Micrococcus, Serratia, Proteus, Salmonella, Klesbsiella, Streptococcus) in various potable water sources in Nigeria [43]. Based on Figure 1, the high microbial isolate similarity suggests resemblance in anthropogenic activities in the river along the sampling locations. Typically, microbes are ubiquitous and therefore known for important roles including decomposition of organic materials, bioaccumulation of chemicals and biogeochemical cycling of elements [42]. Their presence in the sediment of the study area suggests suitable environment for microorganisms to thrive and grow. This suggests that the surface water is not safe for direct consumption and domestic purpose without some treatment [1]. Typically, the presence of microbial isolates, its distribution, abundance and growth is greatly influenced by environmental conditions such as pH, temperature, pressure, availability of nutrients and salinity [42]. Therefore, the variation that exists in the findings of this study compared to previous study on sediment microbial quality from different locations could be associated to the environmental condition and physical and/or chemical nature of the rivers as wells as prevailing anthropogenic activities in such location.
Conclusion
Sediment results from the deposition of organic and inorganic substances from human and natural effect on surface water. Sediment quality appears to be a function of surface water quality. This study investigated the microbial quality of sediment of River Nun at Amassoma axises. The study found that there was significant variation among the various locations. This depicts various levels of anthropogenic activities along the water body. The diversity of microbes found in the sediment suggested possible contamination of the surface water. Therefore, there is the need for treatment of the water prior to domestic use.
References
- Agbabiaka TO, Oyeyiola GP (2012) Microbial assessments of soil sediments of Foma River, Ita-Nmo, Ilorin, Nigeria. International Journal of Applied Biological Research 4: 7-17.
- Seiyaboh, EI, Inyang, IR, Izah, SC (2016a) Spatial variation in physico-chemical characteristics of sediment from Epie Creek, Bayelsa State, Nigeria. Greener Journal of Environment Management and Public Safety 5: 100-105.
- Seiyaboh, EI, Izah SC, Oweibi S (2017a) Physico-chemical characteristics of sediment from Sagbama Creek, Nigeria. Biotechnological Research 3: 25-28.
- Adesuyi AA, Ngwoke MO, Akinola MO, Njoku KL, Jolaoso AO (2016) Assessment of physicochemical characteristics of sediment from Nwaja Creek, Niger Delta, Nigeria. Geoscience and Environment Protection 4: 16-27.
- Ogbonna DN (2014) Distribution of microorganisms in water, soils and sediment from Abattoir wastes in Southern Nigeria. International Journal Current Microbiology and Applied Sciences 3: 1183-1200.
- Ogamba EN, Izah SC, Oribu T (2015a) Water quality and proximate analysis of Eichhornia crassipes from River Nun, Amassoma Axis, Nigeria. Research Journal of Phytomedicine 1: 43-48.
- Agedah EC, Ineyougha ER, Izah SC, Orutugu LA (2015) Enumeration of total heterotrophic bacteria and some physico-chemical characteristics of surface water used for drinking sources in Wilberforce Island, Nigeria. Journal of Environmental Treatment Techniques 3: 28-34.
- Nyananyo BL, Gijo AH, Ogamba EN (2007) The physico-chemistry and distribution of water hyacinth (Eichhornia cressipes) on the river Nun in the Niger Delta. Journal of Applied Science and Environmental Management 11: 133-137.
- Seiyaboh EI, Izah SC, Bokolo JE (2017b) Bacteriological quality of water from river nun at Amassoma Axises, Niger Delta, Nigeria. ASIO Journal of Microbiology, Food Science & Biotechnological Innovations 3: 22-26.
- Seiyaboh EI, Inyang IR, Izah SC (2016b) Seasonal variation of physico-chemical quality of sediment from Ikoli Creek, Niger Delta. International Journal of Innovative Environmental Studies Research 4: 29-34.
- Seiyaboh EI, Izah SC (2017) Bacteriological assessment of a tidal creek receiving slaughterhouse wastes in Bayelsa state, Nigeria. Journal of Advances in Biology and Biotechnology 14: 1-7.
- Ogamba EN, Izah SC, Toikumo BP (2015b) Water quality and levels of lead and mercury in Eichhornia crassipes from a tidal creek receiving abattoir effluent, in the Niger Delta, Nigeria. Continental Journal of Environmental Science 9: 13-25.
- Inengite AK, Oforka NC, Osuji LC (2010) Survey of heavy metals in sediments of Kolo creek in the Niger Delta, Nigeria. African Journal of Environmental Science and Technology 4: 558-566.
- Ogamba EN, Izah SC, Toikumo BP (2015c) Water quality and levels of lead and mercury in Eichhornia crassipes from a tidal creek receiving abattoir effluent, in the Niger Delta, Nigeria. Continental Journal of Environmental Science 9: 13-25.
- Ogamba EN, Ebere N, Izah SC (2017) Heavy metal concentration in water, sediment and tissues of Eichhornia crassipes from Kolo Creek, Niger Delta. Greener Journal of Environment Management and Public Safety 6: 001-005.
- Izonfuo LWA, Bariweni AP (2001) The effect of urban runoff water and human activities on some physico-chemical parameters of the Epie Creek in the Niger Delta. Journal of Applied Sciences and Environmental Management 5: 47-55.
- Ben-Eledo VN, Kigigha LT, Izah SC, Eledo BO (2017a) Bacteriological quality assessment of Epie Creek, Niger Delta Region of Nigeria. International Journal of Ecotoxicology and Ecobiology 2: 102-108.
- Ben-Eledo VN, Kigigha LT, Izah SC, Eledo BO (2017b) Water quality assessment of Epie creek in Yenagoa metropolis, Bayelsa state, Nigeria. Archives of Current Research International 8: 1-24.
- Seiyaboh EI, Izah SC, Oweibi S (2017c) Assessment of Water quality from Sagbama Creek, Niger Delta, Nigeria. Biotechnological Research, 3: 20-24.
- Seiyaboh EI, Jackson F (2017) Level and impact of hydrocarbon in sediment characteristics of Imiringi oil and gas field facilities in the Niger Delta. Proceedings of the 15th International Conference on Environmental Science and Technology Rhodes, Greece, 31 August to 2 September 2017.
- Izah SC, Ohimain EI (2018) Ecological risk assessment of heavy metals in cassava mill effluents contaminated soil in a rural community in the Niger Delta Region of Nigeria. Molecular Soil Biology 9: 1-11.
- Izah SC, Bassey SE, Ohimain EI (2017a) Amino acid and proximate composition of Saccharomyces cerevisiae biomass cultivated in cassava mill effluents. Molecular Microbiology Research 7: 20-29.
- Izah SC, Bassey SE, Ohimain EI (2017b) Geo-accumulation index, enrichment factor and quantification of contamination of heavy metals in soil receiving cassava mill effluents in a rural community in the Niger Delta region of Nigeria. Molecular Soil Biology 8: 7-20.
- Izah SC, Bassey SE, Ohimain EI (2017c) Assessment of heavy metal in cassava mill effluent contaminated soil in a rural community in the Niger Delta region of Nigeria. EC Pharmacology and Toxicology 4: 186-201.
- Izah SC, Bassey SE, Ohimain EI (2017d) Assessment of pollution load indices of heavy metals in cassava mill effluents contaminated soil: A case study of small-scale cassava processing mills in a rural community of the Niger Delta region of Nigeria. Bioscience Methods 8: 1-17.
- Pepper IL, Gerba CP (2005) Environmental Microbiology. A laboratory manual. 2nd ed. Elsevier academic press, Amsterdam.
- Benson HJ (2002) Microbiological Applications: Laboratory Manual in General Microbiology/complete version, 5th ed. McGaraw-Hill, New York.
- Cheesbrough M (2006) District Laboratory Practice in Tropical Countries. Low price Edition part 2. Cambridge press, England.
- Izah SC, Ineyougha ER (2015) A review of the microbial quality of potable water sources in Nigeria. Journal of Advances in Biological and Basic Research 1: 12-19.
- Holt JG, Kneg NR, Sneath PHA, Stanley JT, Williams ST (1994). Bergey’s Manual of Determinative Bacteriology. William and Wilkins Publisher, Maryland. New York.
- Ellis D, Davis S, Alexiou H, Handke R, Bartley R (2007) Descriptions of Medical Fungi. 2nd ed. Printed in Adelaide by Nexus Print Solutions, Underdale, South Australia.
- Kurtzman CP, Fell JW (1998). The Yeasts: A Taxonomic Study. 4th ed. Elsevier Science, Amsterdam, The Netherlands.
- Iwuagwu JO, Ugwuanyi JO (2014) Treatment and valorization of palm oil mill effluent through production of food grade yeast biomass. Journal of Waste Management.
- Okoduwa SIR, Igiri B, Udeh CB, Edenta C, Gauje B (2017) Tannery effluent treatment by yeast species isolates from watermelon. Toxics 5: 6.
- Izah SC, Bassey SE, Ohimain EI (2017e) Changes in the treatment of some physico-chemical properties of cassava mill effluents using Saccharomyces cerevisiae. Toxic 5: 28.
- Izah SC, Bassey SE, Ohimain EI (2017f) Cyanide and macro-nutrients content of Saccharomyces cerevisiae biomass cultured in cassava mill effluents. Int J Microbiol Biotechnol 176-180.
- Izah SC, Bassey SE, Ohimain EI (2017g) Assessment of some selected heavy metals in Saccharomyces cerevisiae biomass produced from cassava mill effluents. EC Microbiology 12: 213-223.
- Ogbeibu AE (2005) Biostatistics. A practical approach to research and data handling. Mindex Publishing Company Limited, Benin-City Nigeria.
- Izah SC, Angaye TCN (2016a) Ecology of human schistosomiasis intermediate host and plant molluscicides used for control: A review. Sky Journal of Biochemistry Research 5: 075- 082.
- Izah SC, Angaye TCN (2016b) Heavy metal concentration in fishes from surface water in Nigeria: Potential sources of pollutants and mitigation measures. Sky Journal of Biochemistry Research 5: 31-47.
- Tijani MN, Balogun, SA, Adeleye MA (2005) Chemical and Microbiological assessment of water and booton-sediment contaminations in Awba lake (UI), Ibadan, SW Nigeria. RMZ Materials and Geoenvironment 52: 123-126.
- Ogbonna DN, Ideriah, TJK (2014) Effect of Abattoir wastewater on the physico-chemical characteristics of soil and sediment in Southern Nigeria. Journal of Scientific Research and Reports 3: 1612-1632.
- Izah SC, Aseiba ER, Orutugu LA (2015) Microbial quality of polythene packaged sliced fruits sold in major markets of Yenagoa Metropolis, Nigeria. Point Journal of Botany and Microbiology Research 1: 30-36.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences