Comparison of the Biological Activity of High- Level Disinfection and Sterilization in Medical Devices and Biomedical Equipment
1Research and Development Department, Technical Director, Alkamedica S.A.S. Villamaria, The Republic of Colombia
2Research and Development Department, Leader of Research and Development Projects, Alkamedica S.A.S. Villamaria, The Republic of Colombia
- Corresponding Author:
- Jhon Henry Galvis García
Research and Development Department
Leader of Research and Development Projects
Alkamedica S.A.S. Villamaria
The Republic of Colombia
Tel: +573128963570
E-mail: liderproyectos@alkamedica.com
Received Date: October 11, 2017; Accepted Date: October 25, 2017; Published Date: October 31, 2017
Citation: Cortés AMC (2017) Comparison of the Biological Activity of High-Level Disinfection and Sterilization in Medical Devices and Biomedical Equipment. J Health Hyg. Vol.1 No.1:5
Copyright: © 2017 Cortés AMC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The need to select suitable disinfectants for disinfection processes in biomedical equipment and in hospital environment has been highlighted for several decades in multiple scientific articles. Dozens of notes documenting infections in patients have also been published after inadequate disinfection procedures which have been applied to the elements used for their care and treatment. Hence, the aim of the investigation was to compare the biological activity of high-level disinfection and sterilization in medical devices and biomedical equipment with different biochemical and molecular analytical techniques (spectrophotometry, in-house PCR, luminometry, etc.). The swabs were taken from medical devices and biomedical equipment treated by High-Level Disinfection (Glutaraldehyde 0.17%, pH=6, without activator) and different methods of sterilization (Steam, Ethylene Oxide, Plasma, Hydrogen Peroxide, etc.) in different institutions of reference in Colombia. Samples were seeded in culture media specific for the growth of microorganisms of the species Bacillus subtilis, Clostridium difficile and Mycobacterium tuberculosis. The genetic identity of the microorganisms was determined, reporting 7 polymorphic variants of Mycobacterium tuberculosis, in addition, bioburden was evidenced in biomedical devices and equipment that were previously submitted to sterilization processes. Samples taken from devices and equipment treated with Glutaraldehyde 0.17% (pH=6, without activator) did not report growth of the microorganisms of interest. Comparisons between sterilization methods (Ethylene Oxide, Formaldehyde, Vapor, etc.), evidences failures in the elimination of both reproductive and vegetative forms of microorganisms of pathological interest, specifically Acinetobacter baumannii, Staphylococcus warneri, Staphylococcus epidermidis, Brevibacterium spp., Pseudomona luteola and Sphingomonas paucimobilis.
Keywords
Bacillus subtilis; Clostridium difficile; Glutaraldehyde; High-Level Disinfection; Sterilization; Mycobacterium tuberculosis
Introduction
Several international organizations, including the Association of Operating Room Nurses [1,2] and the Centers for Disease Control [3], have established guidelines and regulations for instrumental processing based on classifications Spaulding (Critical, Semicritical and Non-critical), based on the occurrence of nosocomial diseases [4]. Critical devices must be subjected to Sterilization (Physical) or High-Level Disinfection (HLD) (Chemical) processes, for which different technologies have been developed in sterilization processes (Steam, Ethylene Oxide, Plasma, Hydrogen Peroxide, etc.) and High-Level Disinfection (HLD) (Formaldehyde, Glutaraldehyde, Ortophthaldehyde, etc.).
HLD is generated by immersing the instruments in a Liquid Chemical Germicide (GQL), for less time than required for sterilization. For example, Glutaraldehyde 2% (Alkaline) achieves HLD in less than 1 hour; however, it may not necessarily inactivate many bacterial spores when used as a disinfectant. Glutaraldehyde 2% is sporicidal during soak times as short as 10 minutes; however, any disinfection or sterilization process may fail if the instruments are not thoroughly cleaned at first [5-7].
The heat and gas of ethylene oxide (EtO) are the two most commonly used sterilization agents for the reprocessing of medical instruments. Pressurized steam is the best option for sterilization because of its reliability, availability and costeffectiveness. However, flexible endoscopes contain components that can be altered by heat, which prevents sterilization by this method [8].
Equipment using EtO gas is used to reprocess flexible endoscopes and other heat-sensitive instruments, but due to this process, at low temperature, needs the instrument to be aerated for 24 hours prior to its reuse, sterilization with EtO is not feasible. Institutes where endoscopies performed usually have a limited supply of these equipment, therefore, cannot tolerate reprocessing times greater than 1 hour [8].
HLD on endoscopic is the global standard [9] because it is quickacting, cost-effective and has not been shown to pose a risk of infection. It is recommended by the Association of Professionals in Infection Control and Epidemiology [10,11], the Centers for Disease Control and Prevention [13], the Association of Nursing Room Surgery [2] and different organizations for the reprocessing of heat-sensitive instruments [14].
HLD is most often achieved by soaking the instruments in a 2% aqueous solution of alkaline Glutaraldehyde [11-15], which does not deteriorate or corrode metals, gums, plastics, optical equipment, or endoscopes [12,16]. During manual reprocessing, the endoscope is usually immersed in a bucket filled with this germicide [17]. This practice may expose the staff and the surrounding environment to the Glutaraldehyde solution and its vapors. Automatic endoscope scanners, which automatically disinfect and rinse the endoscope, are alternatives to manual reprocessing. They are closed systems that reduce the measured levels of glutaraldehyde vapors in the air at levels much lower than those allowed to contain the germicide in internal chambers [8,18].
Hence, glutaraldehyde solutions are more stable at an acidic pH (3.0 to 6.3), but have a lower biocidal activity than the basic solution. The activated glutaraldehyde solution (pH 7.5 - 8.5) is only stable for 14 days, although it is not advisable to use it for more than a week, due to the molecules polymerize at a pH above 8.5, blocking the aldehyde groups responsible for the biocidal activity [8,12,16,18].
Likewise, there is the risk of progressively diluting the solution after immersing in it instrumental with residual moisture. To avoid this phenomenon, the renewal frequency of the solution varies from 24 hours to one week depending on the frequency of use, in addition, it should be checked that the material is dry before being immersed in that activated solution. Glutaraldehyde diluted in water in concentrations of 0.1% to 1.0%, is used as a cold disinfectant of medical and scientific equipment that is heat sensitive, including dialysis and surgical instruments, suction vials, bronchoscopies, endoscopies, and the ear, nose, and throat instruments. Its effectiveness is more limited compared to algae and fungi [19].
The Glutaraldehyde 2% and pH 7.5 - 8.5 solutions are effective against vegetative bacterial forms in less than 2 minutes, specifically against Mycobacterium tuberculosis (not all publications agree on these results), fungi and viruses, less than 10 Minutes. In the case of spores of species of the genus Clostridium and Bacillus the effectiveness is clear in 2 hours. However, resistance has been shown in species of the genus Aspergillus and Mycobacterium, for which longer contact times (10 hours) are needed to behave as sporicidal, i.e. to generate HLD [20].
All products based on aldehydes and derivatives, including those whose purpose is HLD, do not have a solution concentration validation mechanism by acid-base titration methods, i.e. pH verification, since the latter parameter does not allow the determination of the concentration, considering that it is not an analytical method of quantification of the same. In addition, "check strips" (erroneously used for the quantification of the concentration of substances), which in the strict sense are pH verification strips, are not standardized items to determine the concentration in any solution (they are designed for a specific product), which goes against any analytical method of concentration determination [8,18].
Materials and Methods
Study area
The study was carried out in 3 main Health Institutions in Colombia, of which, for reasons of confidentiality, will not divulge information corresponding to the staff members or the name of the institutions that endorsed the investigation. At each site, samples (swabs) were taken, for triplicate, of instruments (processed and unprocessed) of services such as external consultation, hospitalization, surgery and laboratory. The samples were taken following the notation and method of the Standard Methods for the Examination of Water and Wastewater [21], which were transported to the Clinical Laboratory of the Fundación Hospital Universitario Metropolitano and to the Laboratory of Environmental Studies in Water and Soil of the University of Caldas. These samples were conserved under the parameters described in the protocols of APHA et al. [21]. Samples were processed to determine the presence of pathogenic microorganisms of interest in human health.
In situ bioassay
To evaluate the prevalence (presence/absence) of microorganisms, samples (swab) were collected before and after application of the High-Level Disinfectant (Glutaraldehyde 0.17% in solution, pH=6, without activator) and Sterilization process (Formaldehyde, Hydrogen Peroxide), evaluating cleaning and disinfection by Luminometry (SystemSURE Plus) [22,23]. This swab was seeded in culture media specific for the growth of Bacillus (Nutrient Agar), Clostridium (BD Agar for Clostridium difficile supplemented with 7% sheep blood) and Mycobacterium (Löwestein-Jensen Agar and Stonebrick Agar) [24-28]. After the incubation period, microbial growth was determined by turbidity measured by spectrophotometry at 570 nm emission (Countess II FL - Cell Counter Thermo Fisher) [29,30].
DNA extraction, in-house PCR and partial 16S rRNA gene sequencing
Mycobacterial DNA extraction was modified from protocol of Woo et al. [31]. Briefly, inoculum of mycobacterial colonies on Löwenstein-Jensen and Stonebrick medium were washed with 500 mL 0.1 M Tris/HCl (pH 7.5), and the pellet was suspended in 100 mL lysis solution containing 0.1% NaOH and 0.025% SDS. The mixture was incubated at 60°C for 45 min, followed by addition of 100 mL 0.1 M Tris/HCl (pH 7.5). The DNA extract was stored at 220°C before PCR.
In-house PCR for MTB complex and MAC
Each PCR reaction contained 10 mL DNA extract. A manual onetube nested PCR for IS6110 in MTB complex was performed [32-35]. PCR for MAC was performed according to the protocol published by Li et al. [40], with minor modifications. Of the two forward primers, MAC1 (59-GGACCTCAAGACGCATGTCTTCTG-39) was derived from positions 138–161 of the 16S rRNA gene of M. avium, and MAC2 (59-GGACCTTTAGGCGCATGTCTTTAG-39) was derived from positions 128–151 of the M. intracellulare 16S rRNA gene. The design of the reverse primer MACR (59-GCTCTTTACGCCCAGTAATTCCGG-39) was based on a sequence which is common for both species. A 100mL reaction mixture consisted of 10 mM Tris/HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.2 mM dNTP, 0.4 mM each primer, and 2 U AmpliTaq Gold polymerase (Perkin Elmer).
To activate the Taq polymerase, the mixture was first incubated at 94°C for 10 min. The reaction mixture was then subjected to 50 cycles of amplification (94°C for 1 min, 68°C for 1 min and 72°C for 1 min), with a final single extension at 72°C for 10 min. A 10 mL aliquot of PCR product was electrophoresed for 1 h through a 2% agarose gel in 1 x TBE, and the target band of 390 bp was visualized under UV illumination.
Partial 16S rRNA gene sequencing
The broad-range primer 285 (59-GAGAGTTTGATCCTGGCTCAG-39) and the Mycobacteria specific primer 264 (59-TGCACACAGGCCACAAGGGA-39) were used for amplification, corresponding to Escherichia coli 16S rRNA positions 9–30 and 1027–1046, respectively [36]. The size of the amplicon was about 1040 bp, depending on species. PCR was carried out in a 50 mL reaction mixture having 5 mL DNA template, 10 mM Tris/HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.2 mM dNTP, 0.4 mM each primer, and 2 U AmpliTaq Gold polymerase. To activate the Taq polymerase, the mixture was first incubated at 94°C for 10 min.
The reaction mixture was then subjected to 35 cycles of amplification (94°C for 1 min, 65°C for 1 min and 72°C for 2 min), with a final single extension at 72°C for 10 min. A 5 mL aliquot of the PCR product was electrophoresed for 1 h through a 2% agarose gel in 1 x TBE. Subsequently, PCR products were purified using the QIAquick PCR Purification kit (Qiagen) to remove the unpolymerized primers and deoxynucleoside triphosphates before cycle sequencing. Nucleotide sequences from both DNA strands were determined using the same forward primer 285 and reverse primer 259 (59-TTTCACGAACAACGCGACAA-39) corresponding to E. coli 16S rRNA position 590–609 [36]. The purified fragment was subjected to cycle sequencing by the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (version 3.0), at a quarter of the recommended reaction volume, and an ABI 377 Genetic Analyzer (Applied Biosystems).
For all samples, the sequences of both strands of the amplicons were determined. The generated sequences were assembled and edited using the EDITSEQ version 4.0 program in the DNASTAR software (Lasegene) (DEMO), followed by a FASTA (DEMO) search in Ribosomal Differentiation of Medical Micro-organisms (RIDOM) (DEMO). A sequence match of ≥ 99% with that of the prototype strain sequence in a repository was used as the criterion for species, group or complex level identification [33]. A sequence was considered adequate if the edited sequence had 1% or less ambiguity [33]. A reference strain, Mycobacterium smegmatis (ATCC 700084), was run in parallel as control.
Statistical analysis
Data processing was performed using Shapiro-Wilk normality tests, also Factorial Variance Analysis, to prove differences between treatments and Tukey post hoc tests (p<0.05), to verify the maximum variation. Mean and median differences were tested to prove significant differences between the treatments and before and after the cleaning and disinfection procedure (HLD and Sterilization). All analysis was performed in Statgraphics Centurion version 15.01 (DEMO).
Results
The luminometric analysis was performed in different devices and biomedical equipment, ensuring that at this place they did not have a previous process of disinfection. According to Table 1, significant differences (p<0.05) were evident between the data reported at the beginning of the disinfection process, where the results obtained with HLD process (Glutaraldehyde 0.17% in solution, pH=6, without activator) show a marked decrease in luminometric values with removal percentages higher than 50%.
| Service | Surface type | Representation | Sample | Results | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Start | 1Final | 2ANOVA (pValue) | % Average removal | |||||||
| HLD | 3STR. | HLD | STR. | HLD | STR. | |||||
| Surgery | Horizontal | Biomedical equipment, May table, lamps, siphons, among others. | May auxiliary table | 112 | 0 | NA | NC | NC | 100% | NC |
| Sterilization center | Autoclave | 5 | NA | 0 | NC | NC | NC | 100% | ||
| Autoclave H2O2 Plasma | NC | NC | 25 | NC | NC | NC | NC | |||
| Blade Washing Machine | NC | NC | 140 | NC | NC | NC | NC | |||
| Table packing | NC | NC | 89 | NC | NC | NC | NC | |||
| Plasma H2O2 | NC | NC | 32 | NC | NC | NC | NC | |||
| Sterilization center | Geometry 1 | With lockbox | Kelly pin | NC | 290 | NC | NC | NC | NC | NC |
| Harmonic pin | NC | 164 | NC | NC | NC | NC | NC | |||
| Geometry 2 | With depth measurements | Ferguzón's cannula** | NC | 3 | NC | NC | NC | NC | NC | |
| Barter Liposuction** | NC | 730 | NC | NC | NC | NC | NC | |||
| Laparoscopic cannula** | NC | 25 | NC | NC | NC | NC | NC | |||
| Cystoscopy | 13 | 0 | 0 | NC | NC | 100% | 100% | |||
| Equipment A (Endoscopy) | 913 | 19 | 13 | 0,0300282 | 0,0300335 | 97.9% | 98.5% | |||
| Equipment B (Endoscopy) |
669 | 47 | 78 | 0,0194816 | 0,489435 | 92.9% | 88.3% | |||
| Equipment C (Endoscopy) |
176 | 6 | 21 | 0,112955 | 0,0233128 | 96.5% | 88,1% | |||
| Geometry 4 | Various boxes or containers | Tray HLD | 213* | 90 | NA | NC | NC | 57.7% | NC | |
| Enzymatic tray | 78 | 21 | NA | NC | NC | 73.1% | NC | |||
Table 1 Evaluation of biological action of the processes of Sterilization and HLD.
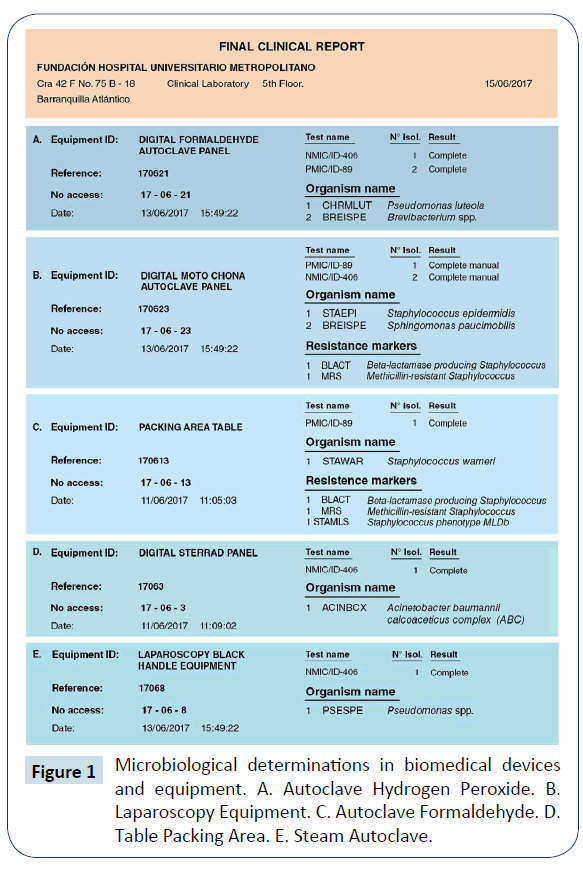
According to the microbiological reports, it was evidenced the presence of different pathogenic microorganisms in the sterilized devices, for example, Acinetobacter baumannii, Staphylococcus warneri, Staphylococcus epidermidis, Pseudomona luteola and Sphingomonas paucimobilis (Figure 1).
Samples of microorganisms that showed growth in solid culture media were sampled to verify their genetic identity, reporting for Mycobacterium spp., 7 spoligotypes of Mycobacterium tuberculosis from biomedical devices and equipment that were previously sterilized by Hydrogen Peroxide, Steam, Plasma and Formaldehyde. Biomedical devices and equipment subjected to HLD (Glutaraldehyde 0.17% in solution, pH=6, without activator) did not report growth in the specific solid culture media for each microorganism of interest (Table 2).
| Service | Surface type | Repr. | Sample | 1baar+(autoclave) | 2baar+(hld: glutaraldehyde 0.17% in solution,Ph = 6, without activator) | 3isolation(hld) | 4isolation(autoclave) | PCR+ | 7no.Spoligotypes | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5L-JUFC/mL | 6S-BUFC/mL | 5L-JUFC/mL | 6S-BUFC/mL | ||||||||
| Surgery | Horizontal | Biomedical equipment, May table, lamps, siphons, among others. | May auxiliary table | 0 | ND | 0 | 0 | ND | ND | 0 | 0 Mtb |
| Sterilization center | Autoclave | 22 | ND | *ND | *ND | 6 | 7 | 48 | 7 Mtb 4 Mtb |
||
| Sterilization center | Geometry 2 | With depth measurements | Cystoscopy | 24 | 0 | 0 | 0 | 1 | 4 | 22 | 1 Mtb 2 Mtb |
| Equipment A (Endoscopy) | 0 | 0 | 0 | 0 | 0 | 2 | 44 | 0 Mtb 2 Mtb |
|||
| Equipment B (Endoscopy) |
1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 Mtb | |||
| Equipment C (Endoscopy) |
1 | 0 | 0 | 0 | 2 | 0 | 44 | 1 Mtb 0 Mtb |
|||
| Geometry 4 | Various boxes or containers | HLD tray | 0 | ND | 0 | 0 | ND | ND | 0 | 0 Mtb | |
| Enzymatic tray | 0 | ND | 0 | 0 | ND | ND | 44 | 1 Mtb 0 Mtb |
|||
Table 2 Mycobacterial determination in samples of biomedical devices and equipment undergoing sterilization and HLD processes.
Discussion
The luminometric results (RLUs) show the presence of organic matter in the samples treated under sterilization procedures, however, this parameter does not allow comparisons about the presence/absence of microorganisms of clinical interest. HLD (Glutaraldehyde 0.17% in solution, pH=6, without activator) has been shown to be an efficient method for the elimination of vegetative forms of microorganisms of clinical interest, for example, Bacillus subtilis, Clostridium difficile and Mycobacterium tuberculosis.
Comparisons between sterilization methods (Ethylene Oxide, Formaldehyde, Vapor, etc.), evidences failures in the elimination of both reproductive and vegetative forms of microorganisms of pathological interest, specifically Acinetobacter baumannii, Staphylococcus warneri, Staphylococcus epidermidis, Brevibacterium spp., Pseudomona luteola and Sphingomonas paucimobilis.
The results obtained agree with data found by Ramírez et al. [37] who, when they were informed of the microbiological results at the General Hospital of zone 32, Mario Madrazo Navarro, Villa Coapa, aimed at evaluating the presence of Acinetobacter baumannii (Gram-negative multi-resistant Bacillus), initiated protocols for the control of the outbreak Mycobacterium tuberculosis Hygiene and washing of hands and equipment with the prepared solution of antimicrobial monoenzymatic detergent (Alkazyme®) and High-Level Disinfectant based on Glutaraldehyde 0.17% in solution, pH=6, without activator (Alkacide®).
According to the research carried out by Tupiza & Vilatuña [38], the recommendations for the High-Level Disinfection process in endoscopy equipment were generated, using Glutaraldehyde 0.17% in solution, pH=6, without activator (Alkacide®), reporting that the product destroys most microorganisms including M. tuberculosis, as found in the present investigation.
In the same way, Santiago [39,40] evaluated different glutaraldehyde-based products, reporting that the Glutaraldehyde 0.17% in solution, pH=6, without activator (Alkacide®) solves the problem of rapid loss of stability and decrease the time of exposure, keeping an excellent antimicrobial activity during a period of 28 to 30 days.
It is important to emphasize that, although biological control is one of the most used indicators, to perform the traceability of the sterilization process, this tool is based on the "ideality" of the process, which means that it does not contemplate the reality of the procedure. For future research, it is recommended the determination of the presence of genes that cause infectious processes of the spoligotypes found of M. tuberculosis.
References
- Association of Operating Room Nurses (1992) Recommended practices. Disinfection. AORN J 56: 715-720.
- Association of Operating Room Nurses (1994) Proposed recommended practices for chemical disinfection. AORN J 60: 463-466.
- Centers for Disease Control-CDC (1985) Recommendations for preventing possible transmission of human T-lymphotropic virus type III/lymphadenopathy-associated virus from tears. MMWR. 34: 533-534.
- Spaulding EH (1968) Chemical disinfection of medical and surgical materials, In: Lawrence CA, Block SS (Eds) Disinfection, Sterilization, and Preservation. Philadelphia, Lea Q Febiger, pp 517-531.
- American Society for Testing and Materials (1994). Standard practice for cleaning and disinfection of flexible fiberoptic and video endoscopes used in the examination of hollow viscera. In: Designation; pp: 854-859.
- Daschner E (1994) Steris System 1 in Germany. Infect Control Hosp Epidemiol 15:295-296.
- Volyes CR, Sanders DL, Simons JE (1995) Steam sterilization of laparoscopic instruments. Surg Endosc 5: 139-141.
- Muscarella LF (1996) High-Level Disinfection or "Sterilization" of Endoscopes.? Control and Hospital Epidemiology 17: 183-187.
- Bond WW (1993) Biological indicators for a liquid chemical sterilizer: A solution to the instrument reprocessing problem? Infect Control Hosp Epidemiol 14: 309-312.
- Rutala WA (1990) APIC guidelines for selection and use of disinfectants. Am J Infect Control 18: 99-117.
- Rutala WA, Clontz EP, Weber DJ (1991) Disinfection practices for endoscopes and other semi-critical items. Infect Control Hosp Epidemiol 12:282-288.
- Rutala WA, Gergen MF, Weber DJ (1993) Sporicidal activity of chemical sterilant used in hospitals. Infect Control Hosp Epidemiol 14:713-718.
- Garner JS, Favero MS (1986) CDC guidelines for the prevention and control of nosocomial infections. Guideline for handwashing and hospital environmental control, 1985. Supersedes guideline for hospital environmental control published in 1981. Am J Infect Control 14: 110-129.
- Martin MA, Reichelderfer M (1994) APIC guidelines for infection prevention and control in flexible endoscopy. Association for Professionals in Infection Control and Epidemiology, Inc. 1991, 1992, and 1993 APIC Guidelines Committee. Am J Infect Control 22: 19-38.
- Babb JR, Bradley CR (1991) The mechanics of endoscope disinfection. J Hosp Infect 18(suppl A): 130-135.
- Dyas A, Das BC (1985) The activity of glutaraldehyde against Clostridium difficile. J Hosp Infect 6: 41-45.
- Omidbakhsh N (2006) A new peroxide-based flexible endoscope-compatible high-level disinfectant. AJIC 34: 571-577.
- Vesley D, Norlien KG, Nelson B (1992) Significant factors in the disinfection and sterilization of flexible endoscopes. Am J Infect Control 20: 291-300.
- Piédrola G (2000) Preventive medicine and public health. (10th edn) Editorial Elsevier España.
- Leveau JY, Bouix M (2002) Technical manual of hygiene, cleaning and disinfection. (1stedn). Madrid Spain . Mundi Press. p 623 .
- APHA, AWWA, WEF (2005) Standard Methods for the Examination of Water and Wastewater. (21st edn) American Public Health Association: Washington.
- Boyce JM, Havill NL, Dumigan DG, Golebiewski M, Balogun O, et al. (2009) Monitoring the Effectiveness of Hospital Cleaning Practices by Use of an Adenosine Triphosphate Bioluminescence Assay. Infect Control Hosp Epidemiol. 30: 678-84.
- Dávila-Ramírez FA, Díaz-Villamil NT, Fajardo-Granados D, Jiménez-Cruz C (2014) Quality of hygiene in surgery rooms by adenosine triphosphate luminometry. Rev Gerenc Polít Health 13: 266-273.
- Lebrun L, Espinasse F, Poveda JD, Vincent-Levy-Frebault V (1992) Evaluation of nonradioactive DNA probes for identification of mycobacteria. J Clin Microbiol 30: 2476-2478.
- Nolte FS, Metchock B, McGowan JE, Edwards A, Okwumabua O, et al. (1993) Direct detection of Mycobacterium tuberculosis in sputum by polymerase chain reaction and DNA hybridization. J Clin Microbiol 31: 1777-1782.
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters 2011-01-05 (v 1.3).
- Madigan MT, Martinko JM, Parker J (2003) In: Pearson Prentice Hall editor Brock: Biology of Microorganisms, Madrid,
- McFarland J (1907) Nephelometer. J Am Med Assoc 14: 1176-1184.
- Sundstrom C, Nilsson K (1976) Establishment and characterization of a human histiocytic lymphoma cell line (U-937). International Journal of Cancer 7: 565-577.
- Haenni M, Ponsin C, Metayer V, Medaille C, Madec J (2012) Veterinary hospital-acquired infections in pets with a ciprofloxacin-resistant CTX-M-15-producing Klebsiella pneumoniae ST15 clone. Journal Antimicrobial Chemotherapy 67: 770-776.
- Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, et al. (2003) Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol 41: 1996-2001.
- Chan CM, Yuen KY, Chan KS, Yam WC, Yim KH, et al. (1996) Single-tube nested PCR in the diagnosis of tuberculosis. J Clin Pathol 49: 290-294.
- Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, et al. (2000) 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 38: 3623-3630.
- Yam WC, Yuen KY, Seto WH (1998) Direct detection of Mycobacterium tuberculosis in respiratory specimens using an automated DNA amplification assay and a single tube nested polymerase chain reaction. Clin Chem Lab Med 36: 597-599.
- Yuen KY, Yam WC, Wong LP, Seto WH (1997) Comparison of two automated DNA amplification systems with a manual one-tube nested PCR assay for diagnosis of pulmonary tuberculosis. J Clin Microbiol 35: 1385-1389.
- Kirschner P, Springer B, Vogel U, Meier A, Wrede A, et al. (1993) Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol 31: 2882-2889.
- Ramírez MLP, Aranza-Aguilar JL, Varela MA, García A, Vélez-Castro G, et al. (2013) Outbreak of nosocomial infection of the lower respiratory tract by Acinetobacter baumannii in an Internal Medicine service of a General Hospital of Mexico City. Med Int Mex 29: 250-256.
- Tupiza MF, Vilatuña MF (2015) evaluation of the cleaning and disinfection process by administrative staff and auxiliary nursing staff in the ICU of Neonatology of H.G.O.I.A., QUITO JUNIO-AUGUST. Grade Work. Central University of Ecuador. p: 112.
- Santiago C (2010) High-level sanitization and disinfection of inhalation equipment and equipment. Revista Mexicana de Enfermería Cardiológica 18: 40-42.
- Li Z, Bai GH, Von Reyn CF, Marino P, Brennan MJ, et al. (1996) Rapid detection of Mycobacterium avium in stool samples from AIDS patients by immunomagnetic PCR. J Clin Microbiol 34: 1903-1907.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences