Characterization and in Plant Detection of Bacteria that Cause Bacterial Panicle Blight of Rice
Temesgen Mulaw1*, Yeshi Wamishe1 and Yulin Jia2
1Rice Research and Extension Center, University of Arkansas, Stuttgart 72160, Germany
2United States Department of Agriculture-Agricultural Research Service (USDA-ARS), Dale Bumpers National Rice Research Center (DB NRRC), Stuttgart, AR 72160, Germany
- *Corresponding Author:
- Temesgen Mulaw
Rice Research and Extension Center
University of Arkansas, Stuttgart 72160, Germany
E-mail: tmulaw@uaex.edu
Received date: January 04, 2018; Accepted date: January 30, 2018; Published date: February 05, 2018
Citation: Mulaw T, Wamishe Y, Jia Y (2018) Characterization and in Plant Detection of Bacteria that Cause Bacterial Panicle Blight of Rice. Plant Pathology Vol 1:3.
Copyright: © 2018 Mulaw T. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Burkholderia glumae (BPB) presumably induces a grain rot symptom of rice that is threatening to rice production in most rice producing states of the USA. The present study was to identify the causal agent of BPB, virulence based on hypersensitive reactions and distribution of the pathogen within a plant. 178 rice panicles samples were analyzed with semi-selective media (CCNT), polymerase chain reaction (PCR) with bacterial DNA gyrase (gyrB) specific markers, and hypersensitive reactions on tobacco leaves. A total of 73 samples out of 178 produced a yellow bacterial colony with similar morphology on CCNT medium suggesting they were bacterial panicle diseases. However, with PCR reactions we only determined that 45 of 73 were due to B. glumae, and the causal agent for the remaining samples was undetermined. Within the 45 samples, 31 highly, 6 moderately, and 5 weakly virulent isolates were grouped based on lesion sizes of the hypersensitive reactions. Pathogenicity variability among the 45 B. glumae detected suggests that different degrees of host resistance exist. To determine the existence of bacteria in different plant tissues, naturally infected plant parts were examined with CCNT media and PCR analysis. B. glumae was again isolated from seeds followed by stems and sheaths from light yellow pigmented CCNT media. In contrast, roots and leaves show no visible yellow pigment on CCNT. Consistent PCR products were produced from the stem, sheath, and seed, but not from the root and leaves. These findings suggest that B. glumae is distributed in the stem, sheath, and seed, and not in the leaf and root. Together this study demonstrated the usefulness of artificial culture media, tobacco reactions, and DNA test with PCR for characterization of BPB, and distribution of bacteria in plants. These findings will help to understand the mechanism of bacteria translocation in plants.
Keywords
Burkholderia glumae; Bacterial anicle blight (Bpb); Hypersensitive reactions; In-plant detection
Introduction
Rice production in the southern United States has a long history of loss to panicle blighting of unknown etiology. The losses caused by bacterial panicle blight (BPB) could be as high as 70%, including reduced yield and poor milling [1]. Significant yield losses from BPB have been experienced in the rice-producing regions of the Southern United States, including Louisiana, Texas and Arkansas in 1996, 1997, 2000, and the most recently, in 2010 [2]. Currently, this disease has affected rice production in many countries of Asia, Africa, South and North America; it is a typical example of the shifting from a minor plant disease to a major disease due to the changes of environmental conditions [3]. The symposiums of BPB often appears during the rice heading stage and is pronounced when rice is grown under high night temperature and frequent rainfalls predisposing rice to diseases outbreak [4].
Rapid detection and accurate identification of pathogens in plant are critical steps to prevent pathogens dissemination. Pathogen identification based on colony morphology or disease symptoms is difficult, time-consuming and unreliable because of the secondary infection by necrotrphic fungi and the similarity among Burkholderia spp. For example, B. glumae, B. plantarii, and B. gladioli were known to infect rice plants causing similar symptoms [5]. Additionally, proliferation of B. glumae and B. plantarii were found to suppress B. gladioli in rice seeds [5]. Interactions among B. glumae, B. gladioli, B. plantarii and other unknown microorganisms often result in different outcomes of crop damage. For example, B. glumae was found to be responsible for the decrease of grain weight, floret sterility, inhibition of seed germination and reduction of stands in rice seedlings dependent on the outcome of the interactions with other bacteria and the environmental factors such as temperature and drought [6,7].
Previous studies have identified abundance of strains of B. glumae including some highly virulent strains that caused 50 to 75% yield reduction [8,9]. Additionally, it was predicted that the B. glumae strains in different rice-production regions have some undefined differences in their genome and virulence [3]. Furuya et al. demonstrated that the extent of virulence of B. glumae strains can be accurately estimated by the use of hypersensitive cell death on tobacco [10]. However, virulence characteristics of B. glumae isolated from rice, and distribution of the causal agent of bacterial panicle blight (BPB) in rice plants have not been clearly demonstrated. Tobacco hypersensitivity is a fast and convenient way to screen bacterial cultures for pathogenicity. It works particularly well for Pseudomonas but can be variable for Xanthomonas and Ralstonia. Some Xanthomonads may require some tweaking of the environmental conditions the tobacco grown in [11,12], and the response may take up to four days [13,14]. Erwinia amylovora and some of the coryneform bacteria will also cause a hypersensitive response. Ralstonia solanacearum cause various results depending on the race. Race 1 results in chlorosis after two days, race 2 induces a typical hypersensitive response in one day and race 3 results in chlorosis after two to eight days [15].
The genetic identity of Burkholderia species has been analyzed by polymerase chain reaction (PCR) using 16S rRNA sequences [16,17]. The discriminatory power of 16S rRNA is too restricted to reveal the detailed phylogenetic relationships among B. plantarii, B. glumae and B. gladioli because of extremely slow rate of evolution of the 16S rRNA gene, it cannot discriminate closely related microorganisms [18]. On the other hand, the genes encoding the β-subunit polypeptide of DNA gyrase (gyrB) estimated to evolve much faster than the 16S rRNA gene that can be used to develop a specific and sensitive detection method [18] to distinguish among Burkholderia species [5]. Therefore, specific primers developed from the gyrB sequences should be reliable for specific detection and identification of B. glumae and B. gladioli in rice materials.
The aims of this study were to 1) isolate and identify the bacterial panicle blight (BPB) pathogen with culture media; 2) verify the causal agent of BPB with PCR; 3) evaluate virulence with tobacco plants; and 4) determine distribution of B. glumae in plants with PCR.
Material and Methods
Isolation and identification of the pathogen
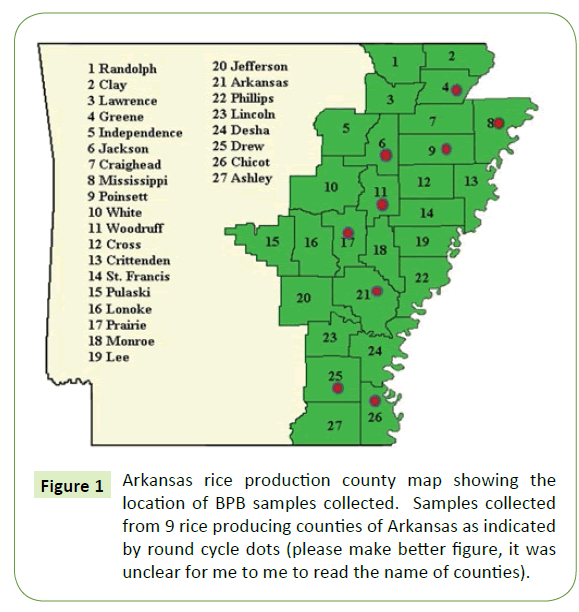
During a 2015 cropping season, 178 naturally infected immature rice panicles with bacterial Panicle blight (BPB) symptoms were collected from growing counties of Arkansas (Supplemental Table S1 and Figure 1). Seeds and florets with discoloration and blanked panicles were collected in paper bags and kept in a refrigerator at 4°C until processing. Seeds were disinfected with 10% sodium hypochlorite for 1 min and rinsed three times with sterile distilled water then left to dry on a sterile filer paper. Disinfected seeds were directly plated on a semi-selective media of CCNT (containing 2 g of yeast extract, 1 g of polypepton, 4 g of inositol, 10 mg of cetrimide, 10 mg of chloramphenicol, 1 mg of novobiocin, 100 mg of chlorotharonil and 18 g of agar in 1000 ml of distilled water, and adjusted to pH 4.8). [19]. From each individual sample 30 seeds were planted on two petri dishes using 15 seeds per dish. Those dishes were sealed using a Para film and incubated at 38°C for 3 to 5 days. The bacterial colonies on these dishes were examined for their morphological characteristics compared with our reference strains of B. glumae. The typical features for bacterial Panicle blight (BPB) on artificial detection media (CCNT) are yellowish white, round, smooth and swollen colonies with a diffusible yellow pigment [19]. Single colonies from each culture plate were collected with a flamed bacteriological loop and streaked on King B medium [20], incubated at 38°C for 48 h, and then stored in Cryo-vial tubes at -80°C in 30% glycerol. Each isolate was given a culture number.
Verification of the causal agent of bacterial panicle blight (BPB) with PCR
The presence of B. glumae was identified with PCR using a pair of primers to detect specific DNA fragments corresponding to the gyrB nucleotide sequences, glu-FW (5’-GAAGTGTCGCCGATGGAG-3’) and glu-RV (5’-CCTTCACCGACAGCACGCAT-3’) [5]. Similar primer pairs of gli-FW (5’-CTGCGCCTGGTGGTGAAG-3’) and gli-RV (5’- CCGTCCCGCTGCGGAATA -3’) were also used to amplify DNA fragments corresponding to the gyrB nucleotide sequences of B. gladioli [5]. PCR amplification was initiated for 20 µl containing 1 µl of template DNA with denaturation at 94°C for 2 min: followed by 35 cycles at 94°C for 1 min, 63°C for 1 min and 72°C for 1 min and 72°C for 10 min as final extension. Aliquots (10 µl) of each PCR products were loaded onto horizontal electrophoresis on a 2% Tris-acetate-EDTA (TAE) agarose gel (Promega) at 80 V for 90 min. Gels were stained with Syber safe for detection of 530 bp, and 479 bp DNA fragments corresponding to the gryB nucleotide sequences of B. glumae and B. gladioli, respectively [5]. A 1-kb ladder (Invitrogen Co.) was used to predict the fragment size of PCR products. However B. plantarii and other Burkholdria spp. are not included with this study because they are not detected as important disease causing pathogen in USA.
Distribution of B. glumae in plants with PCR
To study distribution of B. glumae ten naturally infected rice plants were uprooted from the production fields and brought to a laboratory. Root, stem, sheath, leaf, chuff and seed were collected individually and cleaned with water. These plant parts were disinfected with 1% sodium hypochlorite for 1 min, then rinsed three times with sterile distilled water, and left to dry on a sterile filer paper. Disinfected plant parts were cut to a 1 cm long piece except the seeds that were placed directly on artificial detection (CCNT) media in petri dishes in an incubator at 38°C for 3 to 5 days. DNA was extracted from these plant parts using a DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA), and from bacteria DNA grow on plate media using a UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories, Qiagen, Carlsbad, CA, USA), respectively.
Virulence evaluation with tobacco plants
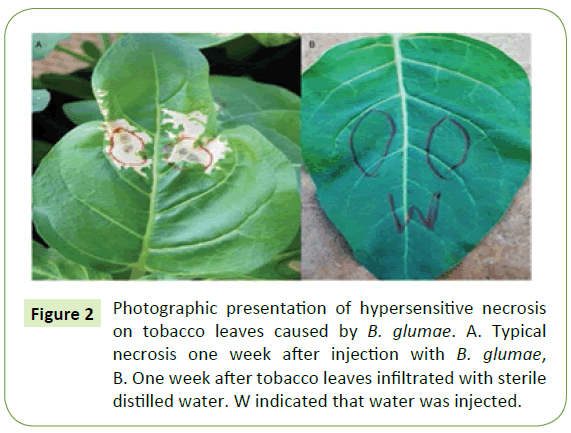
All forty-three isolates identified to be B. glumae were tested for their pathogenicity level with tobacco as described by Furuya et al. [10]. Specifically, tobacco plants (Nicotinaa bethanamiana) were grown to 8 to 9 leaves in approximately 4 weeks after sowing in the greenhouse with a day time temperature ranging between 37°C to 41°C and 75 to 90% relative humidity (RH). Inocula were placed on a King’s B agar (KBA) plates incubated at 38°C for 48 h, then harvested with a sterile cotton swab and suspended in a test tube containing 9 mL of sterile distilled water, and concentration of bacterial suspension were adjusted to be about 108 CFU/mL for inoculation. Three to five tobacco seedlings with the fully expanded leaves were inoculated by injecting at least 3 leaves with 0.5 ml of bacterial suspension using 1 mL sterile syringes and control leaves were injected with sterile distilled water. The control with water did not cause any symptoms one week after injection. The diameters of the lesion of cell death were measured one week after inoculation using four-category disease scale described in Table 1. Large area of necrosis is an indicator for highly virulent strains. After disease scoring, bacteria strains were re-isolated from the diseased tobacco plant to complete Koch’s postulates [21].
| Scalea | Virulentb | Symptoms | No. of isolates |
|---|---|---|---|
| 0 | Not | No symptom produced after inoculation | 0 |
| 1 | Weakly | Slightly browning around the injected site with less than 0.5 cm in diameter | 5 |
| 2 | Moderately | Distinct lesions with 0.5–1 cm in diameter | 9 |
| 3 | Highly | Lesion spreading from the injection with diameter more than 1 cm lesion and even in some isolated cases the whole leaf get wilted completely | 31 |
| aAssigned rating based on necrosis on tabacco leaves one week after injection. bPredicted pathogenicity based on lesion of necrosis on tabacco leaves one week after injection. |
|||
Table 1: Scoring system for tobacco seedling based on the level of hypersensitive reaction to each isolates of B. glumae in greenhouse inoculation tests and number of isolates falling into each category.
Results
Isolation and morphological Identification of the pathogen
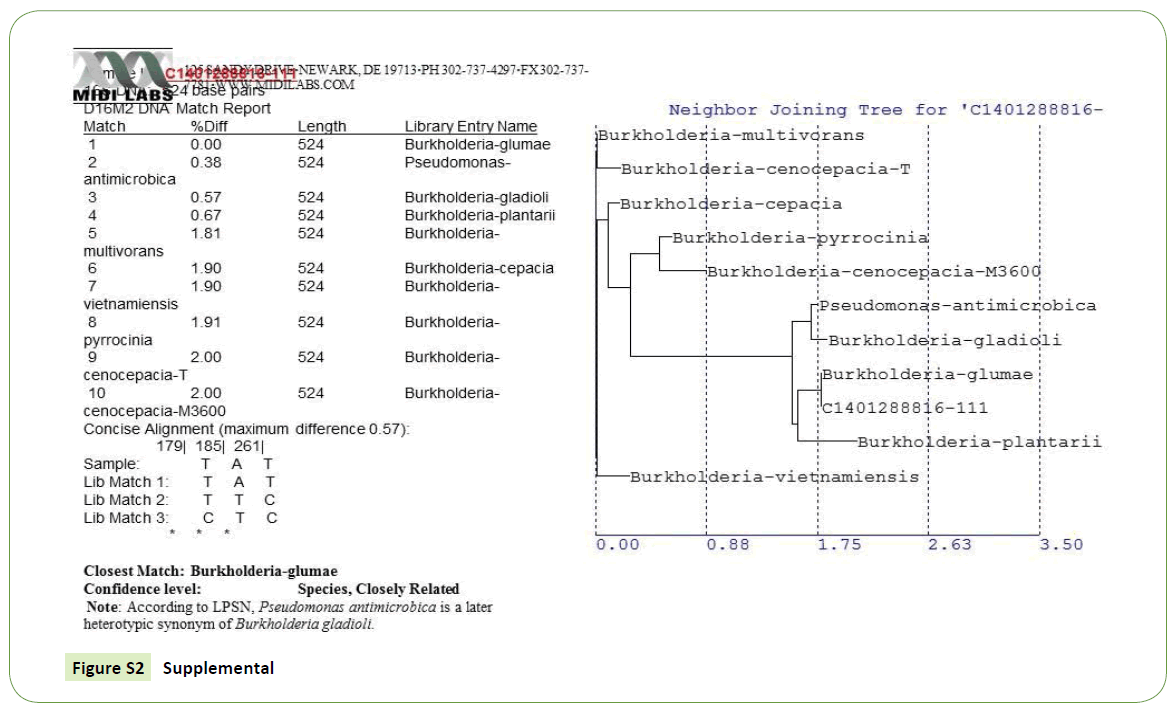
Initial symptoms of the bacterial Panicle blight (BPB) caused by B. glumae were observed on the panicles of the rice plant. Infected panicles with a dark brown discoloration and heavily infected panicles with upright due to blanking were basic characteristics to collect panicle samples from 9 rice producing counties of Arkansas. A total of 178 rice panicle samples were collected (Supplemental Table S1). Seeds and Florets from each collected samples were plated on a semi-selective medium and incubated at a temperature range of 38°C to 40°C for 5 days. The colony characteristics of these samples were compared with our reference strains of B. glumae. About 41% (73 samples) showed similar morphological characteristics to reference strain (Supplemental-2) which is yellowish white, round, smooth and swollen colonies with a diffusible yellow pigment [19] as shown in Table 2. Isolates which have all other morphological characteristics but lacked pigment production also grow well on artificial detection media (CCNT) medium but excluded from this study since it has been reported that this types of strains are not pathogenic to rice [22].
| County | No. of Samples | No. of bacteria Isolated | No. of B. glumae isolates |
|---|---|---|---|
| Praire | 4 | 0 | 0 |
| Lincoln | 13 | 4 | 3 |
| Desha | 3 | 0 | 0 |
| St. Francis | 1 | 1 | 1 |
| Clay | 6 | 3 | 1 |
| Mississippi | 4 | 1 | 1 |
| Craighead | 3 | 2 | 1 |
| Jackson | 11 | 7 | 2 |
| Woodruff | 20 | 0 | 0 |
| Arkansas | 120 | 54 | 34 |
| 178 | 73 | 43 |
Table 2: Total numbers, their morphological and molecular identification of samples with respective of the counties.
Verification of the causal agent of bacterial panicle blight (BPB) with PCR
The identity of seventy-three isolate of bacterial was also confirmed using B. glumae and B. gladioli-specific PCR amplification [5]. An approximately 530 bp DNA fragments of gryB were amplified for 45 isolates indicating that only 62% out of 73 isolates belongs to B. glumae and the remaining twenty-eight isolates did not react with B. glumae-specific primers. On the contrary, no fragments were amplified using B. gladioli-specific primer pairs (Table 3).
| Sample No. | Variety | Virulencea | PCR Ampliconb | |
|---|---|---|---|---|
| glu-FW/glu-RV | gli-FW/gli-RV | |||
| Bg5 | Wells | 3 | Yes | No |
| Bg 6 | Roy J | 3 | Yes | No |
| Bg 7 | 13AR1021 | 3 | Yes | No |
| Bg 14 | CL2134 | 3 | Yes | No |
| Bg 17 | Mermantau | 3 | Yes | No |
| Bg 20 | CLX2008 | 3 | Yes | No |
| Bg 31 | STG12P-23-168 | 1 | Yes | No |
| Bg 32 | RU1401081 | 2 | Yes | No |
| Bg 34 | RU1501133 | 2 | Yes | No |
| Bg 38 | RU1501027 | 3 | Yes | No |
| Bg 41 | STG-12-145 | 3 | Yes | No |
| Bg 44 | RU1501087 | 3 | Yes | No |
| Bg 46 | CL151 | 3 | Yes | No |
| Bg 49 | RU1401161 | 1 | Yes | No |
| Bg 50 | RU1502165 | 3 | Yes | No |
| Bg 53 | RU1403129 | 3 | Yes | No |
| Bg 60 | RU1501093 | 3 | Yes | No |
| Bg 61 | Roy J | 3 | Yes | No |
| Bg 62 | CoDR | 3 | Yes | No |
| Bg 64 | Rex | 3 | Yes | No |
| Bg 73 | RU1501173 | 3 | Yes | No |
| Bg 77 | RU1501133 | 3 | Yes | No |
| Bg 81 | RU1502068 | 3 | Yes | No |
| Bg 87 | RU1504122 | 3 | Yes | No |
| Bg 90 | RU1203190 | 1 | Yes | No |
| Bg 91 | RU1404194 | 3 | Yes | No |
| Bg 108 | RU1301021 | 1 | Yes | No |
| Bg 111 | RU1504186 | 3 | Yes | No |
| Bg 112 | RU1501185 | 3 | Yes | No |
| Bg 113 | RU1303184 | 3 | Yes | No |
| Bg 114 | RU1504193 | 1 | Yes | No |
| Bg 118 | RU1502152 | 3 | Yes | No |
| Bg 119 | RU1501182 | 3 | Yes | No |
| Bg 121 | CL172 | 3 | Yes | No |
| Bg 125 | RU1003113 | 2 | Yes | No |
| Bg 127 | RU1503110 | 3 | Yes | No |
| Bg 128 | RU1502109 | 3 | Yes | No |
| Bg 129 | RU1501148 | 3 | Yes | No |
| Bg 132 | RU1303181 | 2 | Yes | No |
| Bg 135 | RU1501102 | 2 | Yes | No |
| Bg 145 | RU1501108 | 3 | Yes | No |
| Bg 151 | RU1501111 | 2 | Yes | No |
| Bg 153 | CL271 | 2 | Yes | No |
| Bg 155 | CL 111 | 3 | Yes | No |
| Bg 157 | CL 111 | 3 | Yes | No |
| Control | DI Water | H | ||
| a H indicates no visible reaction, 1, weakly virulent, 2, moderately virulent, 3,highly virulent one week after injection respectively. b glu-FW/glu-RV indicates primers specifically to B. glumae and gli-FW/gli-RV indicates primers specifically to B. gladioli, respectively. Yes indicates PCR product produced and No indicates no PCR amplicon. |
||||
Table 3: Results of Virulence level tested by inoculation of Isolates into tobacco leaves to determine pathogenicity level and PCR reaction for two primer sets.
Virulence evaluation with tobacco plants
Reaction to tobacco revealed that all 45 isolates tested are pathogenic at different virulence level (Table 3). About 31 isolates (69%) of the 45 isolates tested were highly virulent (Figure 2A), while nine isolates (20%) moderately virulent. The remaining isolates categorized as weakly virulent isolates whereas plants injected with sterile distilled water remained healthy with no visible hypersensitivity reaction on the leaves (Figure 2B). Koch’s postulates were confirmed by re-isolating from inoculated tobacco leaves and then grow them on a semi-selective media (CCNT) for B. glumae (data not shown) Pathogenic B. glumae isolates produced a yellow pigment, identified as toxoflavin, while non-pathogenic strains did not [22]. Accordingly, all seventy-three isolates tested for their virulence level reislolated from tobacco, and all produced a yellow pigment, which indicated that they are still pathogenic B. glumae bacteria.
Distribution of B. glumae in a plant
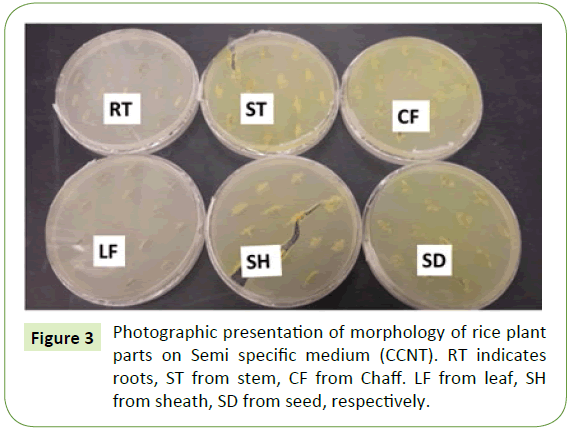
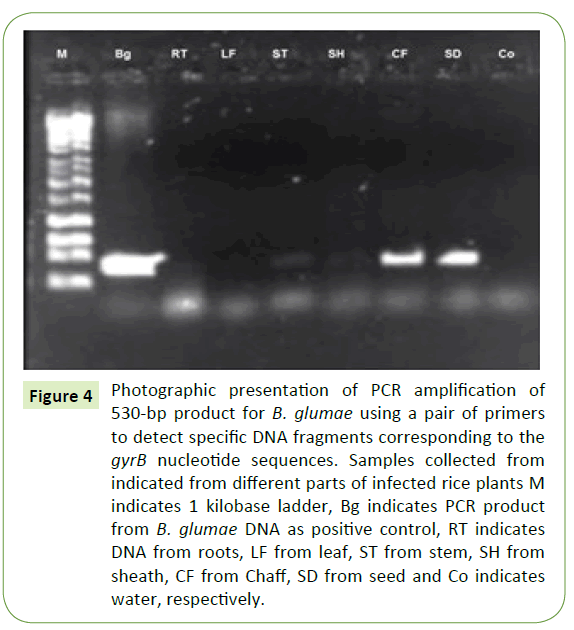
Ten naturally infected rice plants were removed from a rice paddy and different plants were plated on semi-selective media (CCNT). B. glumae were isolated from seed followed by stem and sheath at low concentration level of yellow pigment. However, roots and leaves did not show any visible yellow pigment on semi-selective media (CCNT) (Figure 3). Pathogen identification was confirmed by PCR using B. glumae–specific primer pair with DNA extracted from individual plant parts (root, stem, leaf, sheath, chaff, and seed). PCR products with predicted sizes were obtained from DNA extracted from seed and chaff. No PCR products were amplified from roots and leaves of rice plant but low level of amplification observed for stem and sheath (Figure 4).
Figure 4: Photographic presentation of PCR amplification of 530-bp product for B. glumae using a pair of primers to detect specific DNA fragments corresponding to the gyrB nucleotide sequences. Samples collected from indicated from different parts of infected rice plants M indicates 1 kilobase ladder, Bg indicates PCR product from B. glumae DNA as positive control, RT indicates DNA from roots, LF from leaf, ST from stem, SH from sheath, CF from Chaff, SD from seed and Co indicates water, respectively.
Discussion
Bacterial Panicle blight (BPB) is an emerging bacterial disease that causes significant crop loss worldwide. Characterization of the causal agent for BPB and in plant detection pathogen is an important prerequisite to manage BPB. In the present study, 45 disease samples out of the total 178 from commercial rice fields in the state of Arkansas, USA were determined due to B. glumae. None of the disease samples were caused by B. gladioli suggesting that B. glumae is the causal agent for BPB in Arkansas. The fact that many non B. glumae were isolated from diseased tissue needs further exploration to see if any uncharacterized microorganism can contribute the development of the syndrome and potential ecological relationships with B. glumae. It is well known that the development of disease symptoms and severity of any plant disease not only depends on virulence of the strain, but also on environmental factors, particularly weather conditions. Symptoms typically caused by B. glumae were panicle blighting with floret discoloration (with a gray-brown color), usually on the lower half of the developing grain, with a clear deep brown border followed by sterility or partial filling of the florets causing the panicles to stand erect [23,24]. However, all samples examined in the present study were with these symptoms but some of samples found to be other microorganism but not B. glumae and/or B. gladioli. Our study clearly demonstrated that symptom of bacterial Panicle blight (BPB) was not sufficient to identify the causal agent of this disease.
Apparently, it is challenge to differentiate pathogens that are closely related physiologically and taxonomically by the symptoms they produce and by their growth on selective media. A good identification scheme depends not only on developing a satisfactory resolution level of methods, but also on the group of bacteria studied [25-27]. Semi specific medium (CCNT) is useful for rough screening for bacteria that cause BPB by visualization of unique yellow pigment as indicative of toxoflavin producing bacteria. In the present study we showed that unknown bacteria other than B. glumae and B. gladioli producing similar yellow pigment suggesting that Semi specific medium (CCNT) alone was not sufficient for positive identification of both bacteria. It is fully possible that other unknown bacteria in rice seeds can producing toxoflavin that needs to be further investigated in order to understand their pathogenicity and their bio-control potentials for managing bacterial Panicle blight (BPB) and other rice diseases. In the future, a defined culture medium specifically to B. glumae and B. gladiolia will need to be developed.
We have not found B. gladioli in all diseased samples from Arkansas except B. glumae. To our knowledge, the present study provides the first experimental evidence of B. glumae as the major cause of bacterial Panicle blight (BPB) in Arkansas. This is consistent with that the major causal agent of BPB was B. glumae whereas B. gladioli was less virulent in other geographic regions [28]. In the present study, different isolates of B. glumae show different levels of pathogenicity based on different hypersensitive reaction patterns on tobacco leaves suggesting that there exist genomic and virulence levels variation in Arkansas B. glumae isolates. Forty-five isolates had a hypersensitivity index ranging from weakly to highly sensitive reaction (Table 3). The majority of them caused large necrosis on tobacco suggest that these Arkansas isolates are highly virulent.
In summary, we showed that accurate identification of the causal agent for bacterial Panicle blight (BPB) is challenging, and cross-referencing among two or more detection methods is desirable to ensure that the causal agent can be positively identified. We learned that once you suspect the symptom of rice plant tissue damaged by BPB, the next plausible step is to examine seeds derived from diseased rice plants. If possible, the disease tissues should be obtained from vegetative stage before flowering to localize pathogen in stem and/or sheaths. In contrast, because leaves and roots are not a favorable residences for the B. glumae as compared to seed, stem and sheath. Therefore, it will not be useful to detect pathogen in leaves and roots. Additionally, we demonstrated that there exhibit difference in virulence among B. glumae and these characterized isolates can be used to screen genetic resistance to bacterial Panicle blight (BPB). Together, our findings are useful for plant quarantine and bacterial Panicle blight (BPB) pathogen identification, ultimately these new knowledge will be useful to manage this emerging agronomically important rice disease worldwide.
Note: Isolates that showed yellow pigment on semi-selective media (CCNT) indicated by Yes but if they did not detected using specific primers for B glumae and B. gladioli using PCR they will be indicated with No. That means morphologically similar but not B glumae and B. gladioli because PCR is more specific detection than morphology.
Acknowledgements
The authors thank Scott Belmar, Tibebu Gebremariam of Rice Research and Extension Center, UA, AR and Tracy Bianco of USDA Agriculture Research Service Dale Bumpers National Rice Research Center for their excellent technical supports. This project was supported by Arkansas Rice Research Promotion Board.
References
- Groth DE, Rush MC, Hollier CA (1991) Rice diseases and disorders in Louisiana. Louisiana Agricultural Experiment Station, Louisiana State University Agricultural Center, Baton Rouge, Louisiana: 5-8.
- Ham JH, Melanson RA, Rush MC (2011) Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol 12: 329–39.
- Cui ZB, Xie G, Li B, Huang S (2016) Research Status and Prospect of Burkholderia glumae, the Pathogen Causing Bacterial Panicle Blight. Rice Science 23: 111–118.
- Cha KH, Lee YH, Ko SJ, Park SK, Park IJ (2001) Influence of weather condition at heading period on the development of rice bacterial grain rot caused by Burkholderia glumae. Res Plant Dis 7: 150–154.
- Yukiko M, Hirosuke S, Akinori K, Kouhei O, Naruto F, et al. (2006) Phylogenetic study and multiplex PCR-based detection of Burkholderia plantarii, Burkholderia glumae and Burkholderia gladioli using gyrB and rpoD sequences. Int J Syst Evol Microbiol 56: 1031–1038.
- Miyagawa H (2000) Biocontrol of bacterial seedling blight of rice caused by Burkholderia gladioli using with its avirulent isolate. Japan J Phytopathol 66: 232-238.
- Jeong Y, Kim J, Kim S, Kang Y, Nagamatsu T, et al. (2003) Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis 87: 890–895.
- Francis F, Kim J, Ramaraj T, Farmer A, Rush MC, et al. (2013) Comparative genomic analysis of two Burkholderia glumae strains from different geographic origins reveals a high degree of plasticity in genome structure associated with genomic islands. Mol Genet Genom 288: 195–203.
- Karki HS, Ham JH (2014) The roles of the shikimate pathway genes, aroA and aroB, in virulence, growth and UV tolerance of Burkholderia glumae strain 411gr-6. Mol Plant Pathol 15: 940–947.
- Furuya N, Liyama K, Ueda Y, Matsuyama N (1997) Reaction of Tobacco and Rice Leaf Tissue Infiltrated with Burkholderia glumae or B. gladioli. J Fac Agr Kyushu Univ 42: 43-51.
- Fahy PC, Persley GJ (1983) Plant Bacterial Diseases, a diagnostic guide. Academic Press, New York. 393 pp. Klement, Z. 1982. Hypersensitivity. In Mount, M. S. and Lacy, G. H. (eds). Phytopathogenic Prokaryotes. Academic Press, New York 2: 149-177.
- Lelliott RA, Stead DE (1987) Methods for the diagnosis of bacterial diseases of plants. Oxford, Blackwell Scientific Publications, United kingdom: 216.
- Klement Z (1990) Isolation of bacteria. In: Z Klement, K Rudolph , DC Sands (eds) Methods in Phytobacteriology. Budapest, Akademiai Kiado, Hungary: 95-124.
- Klement Z, Goodman RN (1967) The hypersensitive reaction to infection by bacterial plant pathogens. Annu Rev Phytopathol 5: 17- 44.
- Lozano JC, Sequeira L (1970) Differentiation of races of Pseudomonas solanacearum by a leaf infiltration technique. Phytopathology 60: 833-838.
- Hu FP, Young JM, Triggs CM, Park DC, Saul DJ (2001) Relationships within the Proteobacteria of plant pathogenic Acidovorax species and subspecies, Burkholderia species, and Herbaspirillum rubrisubalbicans by sequence analysis of 16S rDNA, numerical analysis and determinative tests. Antonie van Leeuwenhoek 80: 201–213.
- Salles JF, De Souza FA, van Elsas JD (2002) Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl Environ Microbiol 68: 1595–1603.
- Yamamoto S, Harayama S (1998) Phylogenetic relationships of Pseudomonas putida strains deduced from the nucleotide sequences of gyrB, rpoD and 16S rRNA genes. Int J Syst Bacteriol 48: 813–819.
- Kawaradani M, Okada K, Kusakari S (2000) New selective medium for isolation of Burkholderia glumae from rice seeds. J Gen Plant Pathol 66: 234-237.
- Streeter JG (2007) Factors affecting the survival of Bradyrhizobium applied in liquid cultures to soya bean [Glycine max (L) Merr.] seeds. J Appl Microbiol 103: 1282-1290.
- Félix-Gastélum R, Maldonado-Mendoza IE, Olivas-Peraza NG, Brito-Vega H, Peñuelas-Rubio O, et al. (2017) First Report of Slippery Skin Caused by Burkholderia gladioli in Stored Onion Bulbs in Mexico. Plant Dis 101: 1030
- Wang Z, Yanagita R, Tsuchiya K, Matsuyama N, Wakimoto S (1991) Relationship between pigment productivity and some other bacteriological properties in the mutant strains of Pseudomonas glumae induced by nitrosoguanidine-treatment. Ann Phytopathol Soc Japan 57: 219-224.
- Shahjahan AKM, Rush CM, Clark EC, Groth ED (1998) Bacterial sheath rot and panicle blight of rice in Louisiana. Proc. 27th RTWG 27: 31-32.
- Shahjahan AK, Rush CM, Groth ED, Clark C (2000) Panicle Blight. Rice Science 15: 26-29.
- Stead DE (1992) Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int J Syst Bacteriol 42: 281-295.
- Welch D (1991) Applications of cellular fatty acid analysis. Clin Microbiol Rev 4: 422-438.
- Yang P, Vauterin L, Vancanneyt M, Swings J, Kersters K (1993) Application of fatty acid methyl esters for the taxonomic analysis of the genus Xanthomonas. Syst Appl Microbiol 16: 47-71.
- Nandakumar R, Shahjahan AKM, Yuan XL, Dickstein ER, Groth DE, et al. (2009) Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the southern United States. Plant Dis 93: 896–905.
| Host rice | B.glumae | Species-specific Primer sets | ||||
|---|---|---|---|---|---|---|
| Sample No. | Location/County | Date | variety | Morphological | glu-FW/glu-RV | gli-FW/gli-RV |
| 1 | Praire county | 8/17/2015 | Roy J | No | ||
| 2 | Praire county | 8/17/2015 | CL163 | No | ||
| 3 | Praire county | 8/17/2015 | CLX2134 | No | ||
| 4 | Praire county | 8/17/2015 | Taggart | No | ||
| 5 | Arkansas county | 8/14/2014 | Wells | Yes | Yes | No |
| 6 | Arkansas county | 8/14/2014 | Roy J | Yes | Yes | No |
| 7 | Arkansas county | 8/14/2014 | 13AR1021 | Yes | Yes | No |
| 8 | Arkansas county | 8/13/2014 | CL151 | Yes | No | No |
| 9 | Lincoln county | 8/20/2015 | CL271 | No | ||
| 10 | Lincoln county | 8/20/2015 | CL111 | No | ||
| 11 | Lincoln county | 8/20/2015 | CL151 | No | ||
| 12 | Lincoln county | 8/20/2015 | CL163 | No | ||
| 13 | Lincoln county | 8/20/2015 | RU1301084 | Yes | No | No |
| 14 | Lincoln county | 8/20/2015 | CL2134 | Yes | Yes | No |
| 15 | Lincoln county | 8/20/2015 | CL172 | No | ||
| 16 | Lincoln county | 8/20/2015 | CL151 | No | ||
| 17 | Lincoln county | 8/20/2015 | Mermantau | Yes | Yes | No |
| 18 | Lincoln county | 8/20/2015 | RU1501102 | No | ||
| 19 | Lincoln county | 8/20/2015 | RU1301021 | No | ||
| 20 | Lincoln county | 8/20/2015 | CLX2008 | Yes | Yes | No |
| 21 | Lincoln county | 8/20/2015 | CLX2008 | No | ||
| 22 | Desha County | 8/20/2015 | RU1501105 | No | ||
| 23 | Desha County | 8/20/2015 | Lakast | No | ||
| 24 | Desha County | 8/20/2015 | Roy J | No | ||
| 25 | Arkansas county | 8/24/2015 | STG-04-121 | No | ||
| 26 | Arkansas county | 8/24/2015 | RU1301084 | No | ||
| 27 | Arkansas county | 8/24/2015 | RU1501105 | No | ||
| 28 | Arkansas county | 8/24/2015 | STG-12-145 | No | ||
| 29 | Arkansas county | 8/24/2015 | STG-04-065 | No | ||
| 30 | Arkansas county | 8/24/2015 | RU1501173 | No | ||
| 31 | Arkansas county | 8/24/2015 | STG12P-23-168 | Yes | Yes | No |
| 32 | Arkansas county | 8/24/2015 | RU1401081 | Yes | Yes | No |
| 33 | Arkansas county | 8/24/2015 | RU1501185 | No | ||
| 34 | Arkansas county | 8/24/2015 | RU1501133 | Yes | Yes | No |
| 35 | Arkansas county | 8/24/2015 | Wells | No | ||
| 36 | Arkansas county | 8/24/2015 | RU1401161 | No | ||
| 37 | Arkansas county | 8/24/2015 | RU1501102 | No | ||
| 38 | Arkansas county | 8/24/2015 | RU1501027 | Yes | Yes | No |
| 39 | Arkansas county | 8/24/2015 | RU1501007 | Yes | No | No |
| 40 | Arkansas county | 8/24/2015 | STG-23-168 | No | ||
| 41 | Arkansas county | 8/24/2015 | STG-12-145 | Yes | Yes | No |
| 42 | Arkansas county | 8/24/2015 | RU1501093 | No | ||
| 43 | Arkansas county | 8/24/2015 | STG-06--61 | No | ||
| 44 | Arkansas county | 8/24/2015 | RU1501087 | Yes | Yes | No |
| 45 | Arkansas county | 8/24/2015 | CLX2008 | No | ||
| 46 | St. Francis county | 8/24/2015 | CL151 | Yes | Yes | No |
| 47 | Arkansas county | 8/25/2015 | RU1003123 | No | ||
| 48 | Arkansas county | 8/25/2015 | RU1504198 | No | ||
| 49 | Arkansas county | 8/25/2015 | RU1401161 | Yes | Yes | No |
| 50 | Arkansas county | 8/25/2015 | RU1502165 | Yes | Yes | No |
| 51 | Arkansas county | 8/25/2015 | RU1503169 | No | ||
| 52 | Arkansas county | 8/25/2015 | RU1502128 | Yes | No | No |
| 53 | Arkansas county | 8/25/2015 | RU1403129 | Yes | Yes | No |
| 54 | Arkansas county | 8/25/2015 | RU1501130 | Yes | No | No |
| 55 | Arkansas county | 8/25/2015 | RU0901130 | No | ||
| 56 | Arkansas county | 8/25/2015 | RU1501087 | No | ||
| 57 | Arkansas county | 8/25/2015 | RU1402088 | No | ||
| 58 | Arkansas county | 8/25/2015 | RU1501090 | No | ||
| 59 | Arkansas county | 8/25/2015 | RU1502097 | No | ||
| 60 | Arkansas county | 8/25/2015 | RU1501093 | Yes | Yes | No |
| 61 | Arkansas county | 8/25/2015 | Roy J | Yes | Yes | No |
| 62 | Arkansas county | 8/25/2015 | CoDR | Yes | Yes | No |
| 63 | Arkansas county | 8/25/2015 | RU1505056 | Yes | No | No |
| 64 | Arkansas county | 8/25/2015 | Rex | Yes | Yes | No |
| 65 | Arkansas county | 8/25/2015 | CHNR | No | ||
| 66 | Arkansas county | 8/25/2015 | RU1504083 | Yes | No | No |
| 67 | Arkansas county | 8/25/2015 | RU1301084 | No | ||
| 68 | Arkansas county | 8/25/2015 | RU1501081 | Yes | No | No |
| 69 | Arkansas county | 8/25/2015 | MM14 | No | ||
| 70 | Arkansas county | 8/25/2015 | RU1502131 | Yes | No | No |
| 71 | Arkansas county | 8/25/2015 | RU1502094 | No | ||
| 72 | Arkansas county | 8/25/2015 | RU1503095 | Yes | No | No |
| 73 | Arkansas county | 8/25/2015 | RU1501173 | Yes | Yes | No |
| 74 | Arkansas county | 8/25/2015 | RU1502174 | No | ||
| 75 | Arkansas county | 8/25/2015 | RU1303174 | Yes | No | No |
| 76 | Arkansas county | 8/25/2015 | RU1503132 | No | ||
| 77 | Arkansas county | 8/25/2015 | RU1501133 | Yes | Yes | No |
| 78 | Arkansas county | 8/25/2015 | RU1502134 | No | ||
| 79 | Arkansas county | 8/25/2015 | CL271 | No | ||
| 80 | Arkansas county | 8/25/2015 | RU0901130 | No | ||
| 81 | Arkansas county | 8/25/2015 | RU1502068 | Yes | Yes | No |
| 82 | Arkansas county | 8/25/2015 | RU1303153 | No | ||
| 83 | Arkansas county | 8/25/2015 | RU1502065 | No | ||
| 84 | Arkansas county | 8/25/2015 | RU1503069 | No | ||
| 85 | Arkansas county | 8/25/2015 | RU1401070 | Yes | No | No |
| 86 | Arkansas county | 8/25/2015 | RU1303181 | No | ||
| 87 | Arkansas county | 8/25/2015 | RU1504122 | Yes | Yes | No |
| 88 | Arkansas county | 8/25/2015 | RU1404191 | No | ||
| 89 | Arkansas county | 8/25/2015 | RU1501076 | No | ||
| 90 | Arkansas county | 8/25/2015 | RU1203190 | Yes | Yes | No |
| 91 | Arkansas county | 8/25/2015 | RU1404194 | Yes | Yes | No |
| 92 | Arkansas county | 8/25/2015 | CL151 | Yes | No | No |
| 93 | Arkansas county | 8/25/2015 | RU1502192 | No | ||
| 94 | Arkansas county | 8/25/2015 | RU1502125 | No | ||
| 95 | Arkansas county | 8/25/2015 | RU1504122 | No | ||
| 96 | Arkansas county | 8/25/2015 | RU1503098 | No | ||
| 97 | Arkansas county | 8/25/2015 | Frances | Yes | No | No |
| 98 | Arkansas county | 8/25/2015 | RU1401081 | No | ||
| 99 | Arkansas county | 8/25/2015 | RU1502137 | No | ||
| 100 | Arkansas county | 8/25/2015 | RU1502045 | No | ||
| 101 | Arkansas county | 8/25/2015 | RU1502048 | No | ||
| 102 | Arkansas county | 8/25/2015 | RU0903147 | Yes | No | No |
| 103 | Arkansas county | 8/25/2015 | RU1402051 | No | ||
| 104 | Arkansas county | 8/25/2015 | RU1303181 | No | ||
| 105 | Arkansas county | 8/25/2015 | RU1303153 | No | ||
| 106 | Arkansas county | 8/25/2015 | RU1502031 | Yes | No | No |
| 107 | Arkansas county | 8/25/2015 | RU1304156 | No | ||
| 108 | Arkansas county | 8/25/2015 | RU1301021 | Yes | Yes | No |
| 109 | Arkansas county | 8/25/2015 | RU1404154 | No | ||
| 110 | Arkansas county | 8/25/2015 | RU1402008 | No | ||
| 111 | Arkansas county | 8/26/2015 | RU1504186 | Yes | Yes | No |
| 112 | Arkansas county | 8/26/2015 | RU1501185 | Yes | Yes | No |
| 113 | Arkansas county | 8/26/2015 | RU1303184 | Yes | Yes | No |
| 114 | Arkansas county | 8/26/2015 | RU1504193 | Yes | Yes | No |
| 115 | Arkansas county | 8/26/2015 | RU1502183 | Yes | No | No |
| 116 | Arkansas county | 8/26/2015 | RU1502195 | No | ||
| 117 | Arkansas county | 8/26/2015 | RU1404154 | No | ||
| 118 | Arkansas county | 8/26/2015 | RU1502152 | Yes | Yes | No |
| 119 | Arkansas county | 8/26/2015 | RU1501182 | Yes | Yes | No |
| 120 | Arkansas county | 8/26/2015 | CL163 | No | ||
| 121 | Arkansas county | 8/26/2015 | CL172 | Yes | Yes | No |
| 122 | Arkansas county | 8/26/2015 | RU1502189 | Yes | No | No |
| 123 | Arkansas county | 8/26/2015 | JZMN2 | No | ||
| 124 | Arkansas county | 8/26/2015 | RU1504197 | No | ||
| 125 | Arkansas county | 8/26/2015 | RU1003113 | Yes | Yes | No |
| 126 | Arkansas county | 8/26/2015 | RU1501111 | No | ||
| 127 | Arkansas county | 8/26/2015 | RU1503110 | Yes | Yes | No |
| 128 | Arkansas county | 8/26/2015 | RU1502109 | Yes | Yes | No |
| 129 | Arkansas county | 8/26/2015 | RU1501148 | Yes | Yes | No |
| 130 | Arkansas county | 8/26/2015 | RU1502146 | No | ||
| 131 | Arkansas county | 8/26/2015 | RU1501188 | No | ||
| 132 | Arkansas county | 8/26/2015 | RU1303181 | Yes | Yes | No |
| 133 | Arkansas county | 8/26/2015 | RU1501142 | No | ||
| 134 | Arkansas county | 8/26/2015 | RU1403104 | No | ||
| 135 | Arkansas county | 8/26/2015 | RU1501102 | Yes | Yes | No |
| 136 | Arkansas county | 8/26/2015 | RU1501099 | No | ||
| 137 | Arkansas county | 8/26/2015 | RU1501096 | No | ||
| 138 | Arkansas county | 8/26/2015 | RU1502140 | No | ||
| 139 | Arkansas county | 8/26/2015 | RU1502137 | No | ||
| 140 | Arkansas county | 8/26/2015 | RU1505178 | No | ||
| 141 | Arkansas county | 8/26/2015 | RU1504100 | No | ||
| 142 | Clay county | 9/2/2015 | Antonio | No | ||
| 143 | Clay county | 9/3/2015 | CLX2134 | Yes | No | No |
| 144 | Clay county | 9/4/2015 | CL163 | Yes | No | No |
| 145 | Clay county | 9/5/2015 | RU1501108 | Yes | Yes | No |
| 146 | Clay county | 9/6/2015 | 14SIT891 | No | ||
| 147 | Clay county | 9/7/2015 | RU1501151 | Yes | No | No |
| 148 | Mississippi County | 9/15/2015 | RU1501105 | No | ||
| 149 | Mississippi County | 9/15/2015 | RU15001108 | No | ||
| 150 | Mississippi County | 9/15/2015 | 14SIT891 | No | ||
| 151 | Mississippi County | 9/15/2015 | RU1501111 | Yes | Yes | No |
| 152 | Craighead County | 9/15/2015 | RU1301021 | Yes | No | No |
| 153 | Craighead County | 9/15/2015 | CL271 | Yes | Yes | No |
| 154 | Craighead County | 9/15/2015 | MM14 | No | ||
| 155 | Jackson County | 9/11/2015 | CL 111 | Yes | Yes | No |
| 156 | Jackson County | 9/11/2015 | CL 111 | Yes | No | No |
| 157 | Jackson County | 9/11/2015 | CL 111 | Yes | Yes | No |
| 158 | Jackson County | 10/8/2015 | RU1301084 | No | ||
| 159 | Jackson County | 10/8/2015 | SIT664 | No | ||
| 160 | Jackson County | 10/8/2015 | RU1501130 | No | ||
| 161 | Jackson County | 10/8/2015 | RU1501148 | Yes | No | No |
| 162 | Jackson County | 10/8/2015 | RU1501027 | Yes | No | No |
| 163 | Jackson County | 10/8/2015 | RU1501030 | Yes | No | No |
| 164 | Jackson County | 10/8/2015 | RU1501050 | Yes | No | No |
| 165 | Jackson County | 10/8/2015 | RU 1501027 | No | ||
| 166 | Woodruff County | 7/29/2015 | RTCLXL729 | No | ||
| 167 | Woodruff County | 7/29/2015 | RTXL753 | No | ||
| 168 | Woodruff County | 7/29/2015 | RU1100477 | No | ||
| 169 | Woodruff County | 7/29/2015 | CLX2008 | No | ||
| 170 | Woodruff County | 7/29/2015 | Mermenta | No | ||
| 171 | Woodruff County | 7/29/2015 | RTCLXL745 | No | ||
| 172 | Woodruff County | 7/29/2015 | Taggart | No | ||
| 173 | Woodruff County | 7/29/2015 | RTXP760 | No | ||
| 174 | Woodruff County | 7/29/2015 | RU1301023 | No | ||
| 175 | Woodruff County | 7/29/2015 | CL163 | No | ||
| 176 | Woodruff County | 7/29/2015 | Roy J | No | ||
| 177 | Woodruff County | 7/29/2015 | CL271 | No | ||
| 178 | Woodruff County | 7/29/2015 | Jupiter | No | ||
Supplemental Table-S1: List of samples used in this and results by morphological and conventional polymerase chain reaction assay.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences