Characterisation of Microbial Community in Ballast Water
Lakshmi Eleyadath*, Priya M and Sivanandan Achari V
Department of Environmental Studies, Cochin University of Science and Technology, Kerala, India
- *Corresponding Author:
- Lakshmi Eleyadath
Department of Environmental Studies,
Cochin University of Science and Technology,
Kerala,
India,
Tel: 9495675644;
E-mail: dr.e.lakshmi@gmail.com
Received date: October 06, 2022, Manuscript No. IPWPC-22-14674; Editor assigned date: October 10, 2022, PreQC No. IPWPC-22-14674 (PQ); Reviewed date: October 25, 2022, QC No. IPWPC-22-14674; Revised date: January 10, 2023, Manuscript No. IPWPC-22-14674 (R); Published date: January 18, 2023, DOI: 10.36648/IPWPC.5.1.002
Citation: Eleyadath L, Priya M, Achari SV (2023) Characterisation of Microbial Community in Ballast Water. J Water Pollut Control Vol:5 No:1
Abstract
Majority of the trade around the world is transferred through ships. Buoyancy in ships is maintained by filling in ballast water. Ships during its voyage discharge invertebrates, plants, microorganisms, non-indigenous vertebrates along with the ballast water, creating an ecological imbalance, threatening the naturally evolved biodiversity in the ports. Here, in this study ballast water samples were collected from a ship running in the offshore waters of Indian ocean near Mosel Bay port in South Africa.
Both chemical and microbiological parameters were analyzed for the ballast water samples. On characterization, 5 bacterial strains were isolated. Microbial analysis showed that there is presence of coliform bacteria. From this study alone we cannot conclude that conducting ballast water exchange is not sufficient to meet the D2 standard. More studies must be done to come to conclusion that, with Ballast Water Exchange (BWE), ballast water treatment or with combination of both, D2 standard can be met.
Keywords
Ballast water; Ballast Water Exchange (BWE); Bio-invasion; Characterization; Marine environment
Introduction
Almost two thirds of traded goods worldwide are transported by ship [1]. To ensure ship buoyancy, stability and maneuverability, oceangoing ships need ballast water. Based on estimation, the world seaborne trade in 2013 amounted to 9.35 billion tons of cargo and the global ballast water discharges in 2013 are estimated to about 3.1 billion tonnes [2].
Ballast water in ships contains different invertebrates, nonindigenous vertebrates, phytoplanktons, zooplanktons and microorganisms. One of the major concerns regarding the ballast water is the introduction of these organisms which can be invasive and pathogenic to the new environment [3]. Species transfer by ballast water concerns the spreading of organisms with potentially harmful effects on human health, such as toxin releasing algae or pathogenic bacteria. Marine bio invasion by ballast water threatens the endemic biodiversity creating an ecosystem imbalance. Most of the organisms will not survive longer transit through ballast water, but some smaller organisms like microorganisms survive. Salinity, dissolved oxygen and temperature are the major limiting factors affecting the survival of organisms [4]. If the introduced environment is conducive, organisms will survive and become dominant destroying the endemic organisms. Majority of the ships carry out ballast water exchange for small voyage and it is observed that there is higher probability of organisms being transmitted through small transit. So, characterizing and identifying organisms that are transmitted and discharged to the port of call by the ship is very much important. Introduced species can be lethal to the ecology and socio-economic aspects of the new environment. Here, in this study ballast water samples were collected from a ship running in the offshore waters of Indian ocean near Mosel Bay port in South Africa [5].
Significance of the work
Shipping traffic is one of many human activities in the ocean that has heavy impact on the marine environment. The effects include air pollution, noise pollution, oil spills and spread of invasive species [6]. The ships operating in the offshore sectors undergo ballast water exchange for prevention of bio-invasion through ballast water. During the exchange many organisms, especially invasive species enter the port environment creating an ecological imbalance, competition with the endemic and other already stressed organisms. According to the 2004 convention on ballast water, vessels’ operating in the sea has to comply with the D2 standard. IMO in its latest declaration has notified that by 2024 all the ships operating in the sea has to comply with the D2 standard [7]. So, characterization of ballast water is very much important to understand the effect of ballast water exchange used in the ship for an overall ballast water management. In this paper, we analyze the physicochemical parameters of ballast water and characterization of bacteria from ballast water using biochemical tests and 16S rRNA gene sequencing.
Materials and Methods
Sampling of ballast water
Ballast water samples were collected from a ship running in the offshore sector of Indian ocean waters near Mosel Bay port, South African (Ship is a supply vessel to a nearby rig from the Mosel bay port, cannot reveal the name of the ship). (lat - 34.1747° S, long-22.0834° E). The Indian Ocean occupies an area of 74.92 x 106 km2 including the marginal seas [8]. Its average depth is 3873 m. It is a huge sea area ranging from Eastern Africa to Western Australia and bordered on the North bythe Asian subcontinent. The area between 25° N and 30° S latitude and between 40° E and 98° E longitude. Samples were collected from the ships pump outlet for discharge. Three samples were collected during 18/12/2018 (Sample A), 5/5/2019 (Sample B), 26/12/2019 (Sample C) from the same ship. Grab sampling was done to collect ballast water samples. Samples were transported to the laboratory in a sealed plastic container in an ice box within two days of sampling [9]. Physico-chemical analysis and microbiological analysis were done to the samples.
Physico-chemical analysis
Ballast water samples were analyzed for physico-chemical parameters [10]. pH, dissolved oxygen, conductivity, salinity and chloride parameters were analysed. Analysis was done based on procedures from American Public Health Association (APHAManual on water and waste water quality analysis), 2012.
Morphological characterization of isolated bacteria
To differentiate the bacteria between gram-positive or gramnegative, grams staining was done. Made a thin smear of culture on glass slides, dried the smear and heat fix, covered the smear one by one with crystal violet (60 sec), gramʼs iodine (60 sec), 95% ethanol (20 sec) and safranin (40 sec). Air dried the slides after washing with distilled water and observed under microscope [11].
Biochemical analysis
The biochemical tests was used to determine the ability of the organism to produce catalase test, coagulase, gram staining, urease, indole, methyl red test, Voges Proskauer test, citrate test for identification of isolates [12].
Microbiological analysis
Ballast water samples collected from the ship was used for the isolation and culture of bacteria. Pour plate was done for the characterization of ballast water using sea water agar kept at 37°C incubation for 48 hours. The colonies formed were isolated by streak plate using sea water agar as medium [13]. DNA was extracted from the isolated bacteria colony. The ballast water temperature during sampling was less than 26°C.
DNA extraction, PCR amplification of 16S rRNA sequencing
Cells were procured by centrifugation (12,000 x g for 1 min at 4°C) and chromosomal DNA was isolated from cell pellets. DNA was extracted from the isolated colonies using the genomic DNA kit (origin diagnostics and research) following the spin column protocol for isolation and purification of genomic DNA from cells. Quality and quantity of DNA extracts were estimated using UV spectrophotometer at a wavelength range of 260/280 nm. Optical Density (OD) of DNA extracts were measured and samples showing OD value between 1.7 and 2.0 were selected for PCR thermal amplification [14,15].
After quantification, good quality DNA extracts were stored at 20°C for performing molecular analysis. In order to amplify 16S rRNA gene sequence, PCR amplification kit, master mix (origin diagnostics and research) and 16S rRNA primer pair (27F 5’- AGAGTTTGATCCTGGCTCAG-3’ (Forward primer) and 1514R 5’- AAGGAGGTGATCCAGCC-3’ (reverse primer)) were used. PCR amplification was carried out with a 50 μl reaction mixture using a gradient thermal cycler (Bio-Rad, T-100). The reaction mixture consisted of 5 μl of template DNA (DNA extracts from samples), 2 μl of each primer (forward and reverse), 25 μl of PCR master mix with DNA polymerase (origin) and 16 μl dH2O [16]. The thermal profile comprised of an initial denaturation of the DNA carried out at 95°C for 3 mins, followed by 34 repeats of denaturation at 95°C for 30 secs, annealing at 55°C for 30 secs and extension at 72°C for 30 secs, along with a final elongation carried out at 72°C for 7 mins [17]. PCR products exhibiting intense bands after agarose gel electrophoresis (1.5%) were analyzed to ensure their size with the help of a DNA ladder. These amplicons after gel documentation were selected and outsourced for purification and sequencing [18].
16S rRNA sequence analysis
Manual editing was done on chromatogram of 16S rRNA gene sequences using bio edit 7.0.9 to select evenly spaced peak regions without baselines. Basic Local Alignment Search Tool (BLASTn) was utilized for an initial screening of nucleotide sequences for identifying homologous sequences in public database like NCBI [19]. 16S rRNA sequences were aligned using Clustal X and phylogenetic tree based on Neighbour Joining (NJ) analyses with 1000 bootstraps were generated using MEGA X. In order to confirm the findings of NJ tree, pair wise sequence distance data based on Kimura 2 Parameter model was generated using the same software [20].
Results
General water quality
The physico-chemical characteristics pH, temperature, Eh, dissolved oxygen, total dissolved solids, conductivity and salinity of ballast water were analyzed and recorded (Table 1).
| No. | Parameters | Results | ||
|---|---|---|---|---|
| Sample A (18/12/2018) | Sample B (5/5/2019) | Sample C (26/12/2019) | ||
| 1 | pH | 7.85 | 8.04 | 7.62 |
| 2 | Temperature (oC) | 20 | 22 | 20 |
| 3 | Eh (mV) | -44.1 | -23.8 | -40.1 |
| 4 | Dissolved oxygen (mg/l) | 8.59 | 8 | 8.41 |
| 5 | Total dissolved solids (ppt) | 52.62 | 24.79 | 50.21 |
| 6 | Conductivity (mS) | 56.45 | 26.53 | 54.32 |
| 7 | Salinity (ppt) | 27.54 | 26.92 | 27.32 |
| 8 | E.Coli | Present | Present | Present |
Table 1: Physico-chemical parameters analyzed for ballast water.
Coliform test
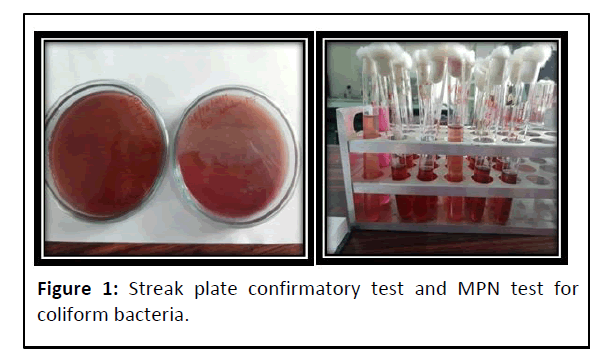
Ballast water samples showed the presence of coliform bacteria in all the samples analyzed (MPN method) and the result was confirmed with the streak plate confirmatory test (Figure 1 and Table 2).
| Sample No. | 24 hrs | 48 hrs | MPN cfu/ml | ||||
|---|---|---|---|---|---|---|---|
| 10 ml | 1 ml | 0.1 | 10 ml | 1.0 cqml | 0.1ml | ||
| Sample A (18/12/2018) | 3 | 1 | 0 | 3 | 3 | 2 | 1100 cfu/ml |
| Sample B (5/5/2019) | 3 | 3 | 2 | 3 | 3 | 2 | 1100 cfu/ml |
| Sample C (26/12/2019) | 3 | 3 | 3 | 3 | 3 | 3 | 1100 cfu/ml |
Table 2: Result of MPN test for coliform bacteria.
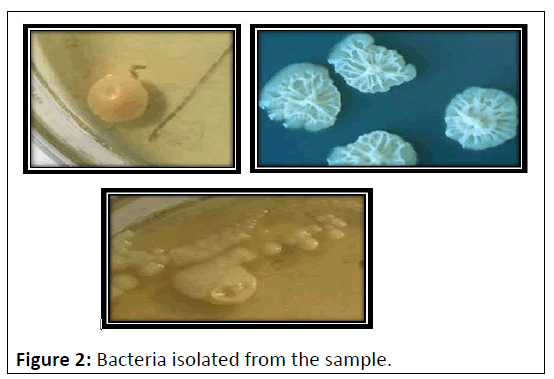
The biochemical tests was used to determine the ability of the organism to produce catalase test, coagulase, gram staining, motility test, nitrate reduction, shape, spore formation, urease, indole, Methyl Red test, voges-proskauer test, citrate test for identification of isolates [21]. From the tests the species identified are Staphylococcus sp, Bacillus sp, Cyto bacillus sp (Table 3 and Figure 2).
| BW 1 | BW 2 | BW 3 | BW 4 | BW 5 | |
|---|---|---|---|---|---|
| Gram staining | +ve | +ve | +ve | +ve | +ve |
| Voges-proskauer test | +ve | +ve | +ve | +ve | -ve |
| Urease test | -ve | -ve | -ve | -ve | -ve |
| Citrate test | +ve | +ve | -ve | +ve | +ve |
| Methyl red | +ve | +ve | +ve | +ve | -ve |
| Indole | -ve | -ve | -ve | -ve | -ve |
| Species identified | Bacillus cereus | Staphylococcus caprae | Cytobacillus kochii | Bacillus licheniformis | Bacillus amyloliquefaciens |
Table 3: Results of Biochemical test.
DNA barcoding, phylogenetic tree and genetic distance data
16S rRNA sequences were successfully generated for bacteria samples cultured from ballast water samples using the mentioned primer pair and thermal profile [22-25]. Homology of these nucleotide sequences with respect to the corresponding species in public database like National Centre for Biotechnology Information (NCBI) were evaluated using BLASTN annotations and they showed more than 99% identity score towards homologous sequences of Bacillus cereus, Bacillus amyloliquefaciens, Bacillus licheniformis, Bacillus kochii (Cytobacillus kochii) and Staphylococcus caprae [26].
Generated 16S rRNA sequences were submitted in National Center for Biotechnology Information (NCBI) database (Table 4). In order to perform molecular analysis, additional homologous 16S rRNA sequence of the above mentioned species were acquired from NCBI and their details are also given in Table 4 [27].
| Sl No. | Species | Status | Accession number |
|---|---|---|---|
| 1 | Bacillus cereus | Generated for this study | MW287217 |
| MW287219 | |||
| Acquired from NCBI | D16266 | ||
| 2 | Bacillus amyloliquefaciens | Generated for this study | MW287224 |
| MW287225 | |||
| MW287228 | |||
| Acquired from NCBI | HE610886 | ||
| 3 | Bacillus licheniformis | Generated for this study | MW287223 |
| Acquired from NCBI | NR_074923 | ||
| 4 | Cytobacillus kochii | Generated for this study | MW287222 |
| Acquired from NCBI | FN995265 | ||
| 5 | Staphylococcus caprae | Generated for this study | MW287221 |
| Acquired from NCBI | NR_024665 |
Table 4: Details regarding 16S rRNA sequences used for molecular analysis.
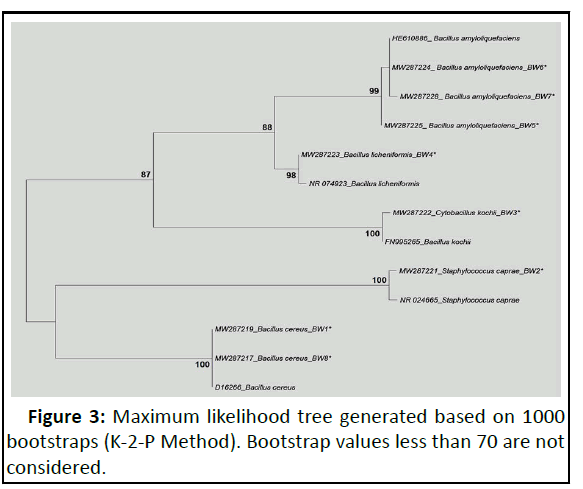
Phylogenetic tree based on Maximum Likelihood (ML), 1000 bootstraps and Kimura 2 parameter and genetic distance data were generated using these sequences (Figure 3).
ML tree exhibited genetic congruency of the incorporated bacteria species as all of the homologous sequences got arrayed with respect to their speciation. From the top, first clade was formed by four sequences of Bacillus amyloliquefaciens within which three sequences (with accession numbers MW287224, MW287225 and MW287228) generated for this study got arrayed with its homologous sequence of B. amyloliquefaciens with accession number HE610886 (acquired from NCBI) with higher bootstrap value (99%). Second clade was formed by two sequences of Bacillus licheniformis within which, one sequence (MW287223) represented the generated one for this study while the second one (with accession number NR_074923), acquired from NCBI [28,29]. Their genetic congruency was evident from its higher bootstrap support (98%). Third clade was also formed by two sequences of Cytobacillus kochii (Bacillus kochii) (with accession numbers MW287222 (generated for this study) and FN995265 (acquired from NCBI) with 100% bootstrap support. Genetic congruency between generated (MW287221) and acquired (NR_024665) sequences of Staphylococcus caprae was witnessed in the fourth clade as they possessed a bootstrap value of 100 [30]. All the members of Bacillus cereus (MW287217, MW287219 and D16266) also exhibited genetic congruency which was evident from their bootstrap value (100%) [31].
Genetic distance data also showed lower intraspecific divergence within the selected species, confirming the findings of ML tree. For Bacillus cereus, there were no genetic distances within the generated sequences as well as the acquired one as the accounted intraspecific divergence was 0. In case of B. amyloliquefaciens intraspecific divergence ranged from 0 to 0.30% [32]. For Bacillus licheniformis and Cytobacillus kochii (Bacillus kochii), the maximum accounted intraspecific divergence was 0.10%. Staphylococcus caprae also exhibited intraspecific divergence up to 0.30%. However, all these intraspecific divergences were lower and contained under the threshold value of 98.6% for delineation of bacterial species [33]. As expected, interspecific divergence was higher for all the selected species. Intraspecific divergences are highlighted in yellow color (Table 5).
| MW287219-Bacillus cereus-BW1* | ||||||||||||
| MW287217-Bacillus cereus-BW8* | 0 | |||||||||||
| D16266-Bacillus cereus | 0 | 0 | ||||||||||
| MW287225-Bacillus amyloliquefaciens-BW5* | 0.073 | 0.073 | 0.073 | |||||||||
| HE610886-Bacillus amyloliquefaciens | 0.074 | 0.074 | 0.074 | 0.001 | ||||||||
| MW287224-Bacillus amyloliquefaciens-BW6* | 0.074 | 0.074 | 0.074 | 0.001 | 0 | |||||||
| MW287228-Bacillus amyloliquefaciens-BW7* | 0.075 | 0.075 | 0.075 | 0.003 | 0.001 | 0.001 | ||||||
| MW287223-Bacillus licheniformis-BW4* | 0.077 | 0.077 | 0.077 | 0.023 | 0.025 | 0.025 | 0.026 | |||||
| NR-074923-Bacillus licheniformis | 0.078 | 0.078 | 0.078 | 0.025 | 0.026 | 0.026 | 0.028 | 0.001 | ||||
| MW287222-Cytobacillus kochii-BW3* | 0.093 | 0.093 | 0.093 | 0.081 | 0.081 | 0.081 | 0.083 | 0.063 | 0.065 | |||
| FN995265-Bacillus kochii | 0.091 | 0.091 | 0.091 | 0.079 | 0.079 | 0.079 | 0.081 | 0.062 | 0.063 | 0.001 | ||
| MW287221-Staphylococcus caprae-BW2* | 0.087 | 0.087 | 0.087 | 0.11 | 0.11 | 0.11 | 0.112 | 0.108 | 0.11 | 0.109 | 0.111 | |
| NR-024665-Staphylococcus caprae | 0.087 | 0.087 | 0.087 | 0.11 | 0.11 | 0.11 | 0.112 | 0.108 | 0.11 | 0.109 | 0.11 | 0.003 |
Table 5: Intraspecific divergence of the species.
Discussion
The general water quality analysis of ballast water shows that the water is alkaline or neutral with dissolved oxygen value ranging from 8.0 mg/l to 8.59 mg/l. Five bacterial species have been identified from both biochemical analysis and 16S rRNA sequencing [34]. Identified species are Bacillus cereus, Staphylococcus caprae, Cytobacillus kochii, Bacillus licheniformis and Bacillus amyloliquefaciens. From the analysis, it is observed that Bacillus is the dominant species identified from the sample.
The identified bacteria Bacillus cereus, Bacillus amyloliquefaciens and Bacillus licheniformis are mostly found in soil and marine waters. Bacillus cereus is found to have close identity with Bacillus thurigiensis whereas Bacillus amyloliquefaciens and Bacillus licheniformis is found to have identity, similar to Bacillus sub ilis [35]. The distinction of bacteria to the one identified from the ballast water sample was confirmed from the biochemical tests and 16S rRNA gene sequencing. Using 16S rRNA sequencing the notable differences in the number and location of prophages, transposable elements were identified to distinguish species from each other.
Initially from sequencing the isolated bacteria, 7 species were identified. Those two species other than 5 identified species were Bacillus sub ilis and Bacillus velenzensis. The species was found to have similarity with Bacillus amyloliquefaciens. On thorough analysis it was found that those isolates were mimicking the identity of Bacillus amyloliquefaciens [36]. The ambiquity in identifying the Bacillus amyloliquefaciens from Bacillus velenzensis and Bacillus sub ilis has been reported by Sacchi et al, 2002. In his study found that one small difference in one base between sequences or partial matches at a single nucleotide position in the 16SrRNA gene is important for species differentiation [37]. They found that the three species Bacillus amyloliquefaciens, Bacillus Velenzensis and Bacillus sub ilis are different species and 16S rRNA has the potential to differentiate strains at the sub species level.
Within the selected species, the genetic distance data showed lower intraspecific divergence, which is confirmed by the ML tree findings. The intraspecific divergences were lower and contained under the threshold value of 98.6% for delineation whereas interspecific divergence was higher for the species identified [38].
The ballast water sample collected during sampling was having less than 26°C temperatures. The culturing and isolation of the colonies were done using sea water agar and was incubated at 37°C [39]. The low percentage of bacteria count from the sample can be corroborated to the difference inincubation temperature during culturing to that of the ballast water sample temperature [40].
The Ship vessel from which sample was collected, perform ballast water exchange to prevent bio invasion. All the species identified from the sample are within the 50 μm size (0.25 μm to 15 μm) [41]. According to the 2004 convention on ballast water, the vessels should meet the D2 standard that is, presence of less than 10 viable organisms with less than 50 μm size.
Quantitative estimation of E. coli was done using MPN method (Table 2). From the analysis, it is observed that all 3 samples had the presence of coliform in the range of 1100 cfu per 100 ml. According to the IMO regulations the group of specified indicator bacteria should be: <1 colony forming unit 100.ml-1 of Vibrio cholera; <250 cfu.100 ml-1 of Escherichia coli and <100 cfu·100 ml-1 of intestinal Enterococci during discharge of ballast water. IMO regulation D-2, ‘ballast water performance standard’ states that indicator microbes in water discharged by ships shall not exceed specified concentrations. These include, but are not limited to: Vibrio cholera, Escherichia coli and intestinal enterococci.
Conclusion
From the ballast water sample analysis 5 bacterial species were identified. Bacillus cereus strain, Staphylococcus caprae strain, Bacillus Kochii strain CN20-4, Bacillus licheniformis strain TB 212, Bacillus amyloliquefaciens strain. The analysis shows the presence of coliform bacteria. Depending on the bacterial profile we can understand the history of ballast water. The study shows that the Ballast Water Exchange (BWE) has not restricted the invasion or presence of bacteria in the ballast water while discharging. This study alone does not validate that the BWE is not an appropriate method for ballast water management. More analysis of samples from ships running in different sea waters and ports needs to be done to come to conclusion that whether BWE, some other ballast water treatment method or a combination of both is appropriate to meet the D2 standard for ships with short voyages.
Role of Funding Source
There is no funding source available for the study.
Conflicts of Interest
There is no conflict of interest
Acknowledgements
We would like to acknowledge Cochin university of science and technology for giving us an opportunity to work on ballast water as post-doctoral fellows.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al. (1995) Short protocols in molecular biology. Trends Biotechnol 10:456
- Baldin RP (1992) Cargo vessel ballast water as a vector for the spread of toxic phytoplankton species to New Zealand. J R Soc 22:229-242
- Carlton JT, Geller JB (1993) Ecological roulette: The global transport of nonindigenous marine organisms. Science 261:78-82
- Chen ML, Tsen HY (2002) Discrimination of Bacillus cereus and Bacillus thuringiensis with 16S rRNA and gyrB gene based PCR primers and sequencing of their annealing sites. J Appl Microbiol 92:912-919
[Crossref] [Google Scholar] [PubMed]
- Christian RR, Pipes WO (1983) Frequency distribution of coliforms in water distribution systems. Appl Environ Microbiol 45:603-609
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, et al. (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial Tag-Encoded FLX Amplicon Pyrosequencing (bTEFAP). BMC Microbiology 8:125
[Crossref] [Google Scholar] [PubMed]
- Doblin MA, Popels LC, Coyne KJ, Hutchins DA, Cary SC, et al. (2004) Transport of the harmful bloom alga Aureococcus anophagefferens by oceangoing ships and coastal boats. Appl Environ Microbiol 70:6495-6500
[Crossref] [Google Scholar] [PubMed]
- Drake JM, Lodge DM (2004) Glocal hot spots of biological invasions: Evaluating option for ballast-water management. Proc Biol Sci 6: 575-580
[Crossref] [Google Scholar] [PubMed]
- Gupta SR, Singbal SYS (1988) Marine pollution in the indian ocean-problems,prospects and perspectives. Indian J Fish 18:333-356
- Hallegraeff GM, Bolch CJ (1991) Transport of toxic dinoflagellate cysts via ship’s ballast water. Marine pollution bulletin 22:27-30
- Hallmich C, Gehr R (2010) Effect of pre and post-UV disinfection conditions on photoreactivation of fecal coliforms in wastewater effluents. Water Res 44:2885-2893
[Crossref] [Google Scholar] [PubMed]
- Haneef UR, Siddique NN, Aman A, Nawaz MA, Baloch H, et al. (2015) Morphological and molecular based identification of pectinase producing Bacillus licheniformis from rotten vegetable. J Genet Eng Biotechnol 13:139-144
[Crossref] [Google Scholar] [PubMed]
- Sieden JM, Way C, Rivkin RB (2010) Microbial hitchhikers: Dynamics of bacterial populations in ballast water during a trans-pacific voyage of a bulk carrier. Aquatic Invasions 5:13-22
- Joachimsthal EL, Ivanov V, Tay ST, Tay JH (2004) Bacteriological examination of ballast water in Singapore harbour by flow cytometry with FISH. Mar Pollut Bull 49:334-343
[Crossref] [Google Scholar] [PubMed]
- Joachimsthal EL, Ivanov V, Tay ST-L, Tay J-H (2004) Bacteriological examination of ballast water in Singapore habour by flow cytometry with fish. Mar Pollut Bull 49:334-343
[Crossref] [Google Scholar] [PubMed]
- Jose D Harikrishnan M (2016) Non-homologous COI barcode regions: A serious concern in decapod molecular taxonomy. Mitochondrial DNA A DNA Mapp Seq Anal 28:482-492
[Crossref] [Google Scholar] [PubMed]
- Kamar R, Gohar M, Jehanno I, Rejasse A, Kallassy M, et al. (2013) Pathogenic potential of Bacillus cereus strains as revealed by phenotypic analysis. J Clin Microbiol 51:320-323
[Crossref] [Google Scholar] [PubMed]
- Khan MS, Gao J, Chen X, Zhang M, Yang F, et al. (2020) The endophytic bacteria Bacillus velezensis Lle-9, isolated from Lilium leucanthum, harbors antifungal activity and plant growth-promoting effects. J Microbiol Biotechnol 30:668-680
[Crossref] [Google Scholar] [PubMed]
- Ki JS, Zhang W, Qian PY (2009) Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. J Microbiol Methods 77:48-57
[Crossref] [Google Scholar] [PubMed]
- Kumar S, Stecher G, Li M, Knyaz C (2018) MEGA X: Molecular evolutionary genetic analysis across computing platforms. Mol Biol Evol 35:1547-1549
- Kumbar B, Mahmood R (2017) Predictive models of 16s rRNA secondary structure from Bacillus subtilis isolates. Biosci Discv 8:776-782
- Lalucat J, Mulet M, Gomila M, Valdes EG (2020) Genomics in bacterial taxonomy: Impact on genus Pseudomonas. Genes 11:139
[Crossref] [Google Scholar] [PubMed]
- Lane N (2011) Energetics and genetics across the prokaryote eukaryote divide. Biol Direct 6:35
- Le TH, Sivachidambaram V, Yi X, Li X, Zhou Z (2014) Quantification of polyketide synthase genes in tropical urban soils using real-time PCR. J Microbiol Methods 106:135-142
[Crossref] [Google Scholar] [PubMed]
- Lilly EL, Kulis P, Gentien P, Anderson DM (2002) Paralytic shellfish poisoning toxins in France linked to a human-introduced strain of Alexandrium catenella from the western Pacific: Evidence from DNA and toxin analysis. J Plankton Res 24:443-452
- Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, et al. (2012) A drought resistance promoting microbiome is selected by root system under desert farming. PLOS One 7:48479
- McCarthy SA, Khambaty FM (1994) International diseemination of epidemic Vibrio cholerae by cargo ship ballast and other non potable waters. Appl Environ Microbiol 60:2597-2601
[Crossref] [Google Scholar] [PubMed]
- McPherson K (1988) Maritime passenger traffic in the Indian ocean region before the nineteenth century. The great circle. 10:49-61.
- Mimura H, Katakura R, Ishida H (2005) Changes of microbial populations in a ship's ballast water and sediments on a voyage from Japan to Qatar. Mar Pollut Bull 50:751-757
[Crossref] [Google Scholar] [PubMed]
- Nel S, Davis SB, Endo A, Dicks LMT (2019) Differentiation between Bacillus amyloliquefaciens and Bacillus subtilis isolated from a South African sugar cane processing factory using ARDRA and rpoB gene sequencing. Arch Microbiol 201:1453-1457
[Crossref] [Google Scholar] [PubMed]
- Pruden A, Pei R, Storteboom H, Carlson K (2006) Antibiotic resistance genes as emerging contaminants: Studies in Northern Colorado. Environ Sci Technol 40 23:7445-7450
[Crossref] [Google Scholar] [PubMed]
- Rey MW, Ramaiya P, Nelson BA, Karpin SDB, Zaretsky EJ, et al. (2004) Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol 5:77
[Crossref] [Google Scholar] [PubMed]
- Ruiz GM, Rawling TK, Dobbs FC, Drake LA, Mullady T, et al. (2000) Global spread of microorganisms by ships. Nature 408:49-50
[Crossref] [Google Scholar] [PubMed]
- Sabir JSM, Abo-Aba SEM, Sabry A, Hussein RM, Bahieldin A, et al. (2013) Isolation, identification and comparartive analysis of 16S rRNA of Bacillus subtilis grown around Rhazya stricta roots. Life Sci 10:980-986
- Sacchi CT, Whitney AM, Mayer LW, Morey R, Steigerwalt A, et al. (2002) Sequencing of 16S rRNA Gene: A rapid tool for identification of Bacillus anthracis. Emerg Infect Dis 8:1117-1123
[Crossref] [Google Scholar] [PubMed]
- Seiler H, Schmidt V, Wenning M, Scherer S (2012) Bacillus Kochii sp.nov, isolated from foods and a pharmaceutical manufacturing site. Int J Syst Evol Microbiol 62:1092-1097
[Crossref] [Google Scholar] [PubMed]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F (1997) The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876-4882
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, et al. (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40:115
[Crossref] [Google Scholar] [PubMed]
- Vary PS, Biedendieck R, Fuerch T, Meinhardt F, Rohde M, et al. (2007) Bacillus megaterium-from simple soil bacterium to industrial protein production host. Appl Microbiol Biotechnol 76:957-967
[Crossref] [Google Scholar] [PubMed]
- Watanabe S, Aiba Y, Tan Xin-Ee, Li FY, Boonsiri T, et al. (2018) Complete genome sequencing of three human clinical isolates of Staphylococcus caprae reveals virulence factors similar to those of Staphylococcus epidermis and Staphylococcus capitis. BMC Genomics 19:810
- Zhenga XW, Yanb Z, Nouta MJR, Smida EJ, Zwieteringa MH, et al. (2014) Microbiota dynamics related to environmental conditions during the fermentative production of Fen-Daqu, a Chinese industrial fermentation starter. Int J Food Microbiol 182-183:57-62
[Crossref] [Google Scholar] [Indexed]
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences