Bioinformatics Analysis of Genes and microRNAs Located between TMPRSS2 and ERG in Prostate Cancer
Song C1*, Ru G1, Guo J1 and Chen H2
1 Medical Research Center, Shaoxing People’s Hospital, Shaoxing, Zhejiang, P.R. China
2 Zhejiang Institute of Microbiology, Hangzhou, Zhejiang, P.R. China
- *Corresponding Author:
- Chunjiao Song
Medical Research Center, Shaoxing People’s Hospital, Shaoxing, Zhejiang, P.R. China.
Tel: 86057588228687
E-mail: chunjiaosong@163.com
Received Date: September 11, 2018; Accepted Date: September 12, 2018; Published Date: September 17, 2018
Citation: Song C, Ru G, Guo J, Chen H (2018) Bioinformatics Analysis of Genes and microRNAs Located between TMPRSS2 and ERG in Prostate Cancer. J Mol Cell Biochem Vol.2 No.2:8
Abstract
Co-relation between diabetes and tuberculosis (TB) creates dual burden of this disease worldwide. This bi directional association necessitates special attention and thus detailed study on human immune system regarding this is necessary. It is very important to develop some good strategies for prevention of diabetes in patients who are exposed to Mycobacterium tuberculosis (M. tuberculosis). Both these diseases are negatively affecting each other as individual with tuberculosis may frequently attain hyperglycaemic condition in diabetic patients and on the other side anti diabetic drugs may also interact with anti tuberculosis drugs in many ways to decreases their efficacy. This review provides recent estimates of TB and diabetes incidence and mortality in the world. In this review, we summarize the current clinical and immunological studies on co-relation between diabetes and TB and what can be done for prevention of these diseases, and spread awareness of this unfavourable co-relation between communicable and non communicable disease which can cause serious threat to public health.
Keywords
Mycobacterium tuberculosis; Alveolar macrophages; Pathogenesis; Diabetes mellitus; Phagolysosome
Abbreviations
TB: Tuberculosis; M. tuberculosis: Mycobacterium tuberculosis; DM: Diabetes Mellitus WHO: World Health Organization; IFN-γ: Interferon γ; IL: Interleukin; TNF-α: Tumour Necrosis Factor-α
Introduction
Prostate cancer (PCa) is one of the most important malignant tumors of the male reproductive system. It is the most frequent malignancy in Europe and America, accounting for 19% of all neoplasms and remains second (8% of male cancer-related deaths) in America [1]. In 2017, 161,360 new cases, and 26,730 deaths from PCa were recorded [1]. In China, the incidence of PCa has been on a rising trend, along with population aging, urbanization, westernization of diets, and advances in detection technologies. The incidence rate was elevated at the rate of 197.54%, from 4.48/100,000 in 1990 to 13.33/100,000 in 2013; while the mortality rate rose at the rate of 46.9%, from 2.26/100,000 to 3.32/100,000 [2]. Moreover, PCa caused an obvious increase in the disease burden, with an increase of 1.03% in disability adjusted life year (DALY). The years of life lost (YLL) due to premature death and disability presented an ascending trend along with age [2]. PCa affects both the quality of life and life expectancy of men, becoming an important topic for the early prevention and treatment of malignant neoplasms.
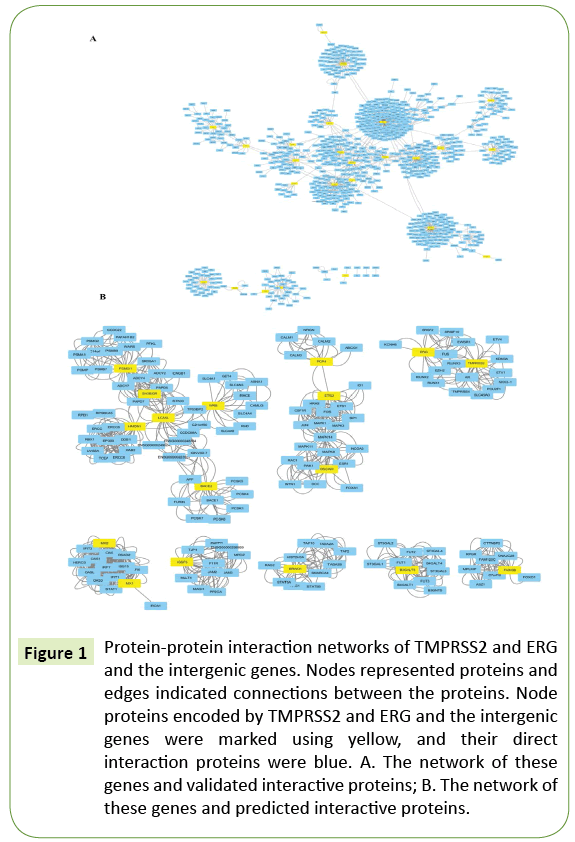
In 2005, Tomlins group initially discovered and reported that ERG (ETS (erythroblast transformation-specific) -related gene) and TMPRSS2 (encoding transmembrane protease, serine 2, a type II trans membrane serine protease regulated by androgen) genes specifically fused in PCa [3]. Since then, TMPRSS2 -ERG fusion gene related studies had become the hotspot of PCa research, including the fusion frequency in PCa patients from different ethnic groups, comparison of detection methodologies, correlation between TMPRSS2 -ERG gene fusion with clinical features, TMPRSS2 -ERG fusion protein signaling pathway, therapeutic methods for ERG overexpression, and so on. The frequency of TMPRSS2 -ERG fusion gene is even higher in younger patients (≤55 years, 63.9%) compared to older patients (≥ 67 years, 40.8%) [4,5]. In a study of 11,152 PCa cases, the data of next generation sequencing (NGS) exhibited that young patients had higher frequency of TMPRSS2 -ERG gene fusion, while older patients were more prone to chromosome deletion. This in turn could be attributed to more potent androgen signaling pathways in younger patients than in older patients. Moreover, high expression of ERG was inversely correlated with the tumor size [6]. Additionally, the frequency of TMPRSS2 -ERG gene fusion was 20-30% in intraductal PCa and 50% in carcinoma in situ, and was further declined in malignant PCa [7-9]. These data indicated that TMPRSS2 -ERG fusion was an early event of PCa, a potentially driven factor to trigger the occurrence of PCa (Figure 1).
Figure 1: Protein-protein interaction networks of TMPRSS2 and ERG and the intergenic genes. Nodes represented proteins and edges indicated connections between the proteins. Node proteins encoded by TMPRSS2 and ERG and the intergenic genes were marked using yellow, and their direct interaction proteins were blue. A. The network of these genes and validated interactive proteins; B. The network of these genes and predicted interactive proteins.
TMPRSS2-ERG fusion gene was specifically expressed in PCa, showing different incidence rates in different ethnic populations. The frequency of TMPRSS2 -ERG fusion was 50% in Caucasian Americans, 31% in African Americans [10], and 18.5% in Chinese population [11]. Combining these reports from multiple nations, the frequency of TMPRSS2 -ERG fusion ranged from 42% to 60% in Europe and America and about 20% in Asia [4,12,13].
TMPRSS2-ERG gene fusion could be generated by chromosome translocation, or chromosome deletion. The frequency of translocation, deletion, and concurrence of the fusion gene was 61.9%, 38.1%, and 0%, respectively in PCa patients with Caucasian American background, while 20%, 60%, and 20%, and respectively with African American background [10]. In 2014, Dong and colleagues evaluated the status of TMPRSS2 -ERG gene fusion in 109 PCa cases (needle core biopsy for 91 cases and radical prostatectomy for 18 cases) from Eastern China. Results revealed that the frequency of TMPRSS2 -ERG gene fusion was 14.3% in tissues of needle core biopsy and 11.1% in tissues of radical prostatectomy. Besides, 13 cases (86.7%) had deletion-type aberration, 7 cases (46.6%) had translocationtype aberration, and 5 cases had the concurrent aberrations of deletion and translocation. Further analysis revealed that 5 cases (38.5%) with metastasis had chromosome deletion; one case with metastasis had the concurrent aberrations, whereas on metastasis occurred in patients with translocation aberration. In this regard, the frequency of TMPRSS2 -ERG gene fusion was explicitly lower in PCa patients from developed Eastern region of China than from Europe and America, and there were more deletion-type. The probability of metastasis was markedly higher in deletion-type than in translocation-type [14]. In 2016, on the basis of PTEN knock-out mouse model, Linn and colleagues constructed two mouse models with TMPRSS2 -ERG gene fusion through translocation or deletion, and functional analysis demonstrated that only deletion-type model might develop into prostatic adenocarcinoma, with poor differentiation and EMT (epithelial mesenchyme transition) feature [15].
In the post-genomic era, bulky genomic data and data mining technologies in life sciences provided methodologies for systematically understanding the complex regulatory networks. Utilizing bioinformatics analysis, the study explored the role and mechanism of the intragenic genes and microRNAs in the progression of PCa, which were deleted when TMPRSS2 and ERG genes fused. The protein-protein interaction (PPI) network diagrams were plotted based on PPI data extracted from several databases, and drew the target gene networks of microRNAs. Subsequently, each nodal protein of the networks was analyzed based on gene ontology (GO) enrichment and signaling pathways. At last, the cellular ultrastructure’s of TMPRSS2 -ERG positive sub line cells and the parent line cells were photographed by transmission scanning electron microscope.
Materials and Methods
Protein-protein interaction network diagram
PPI networks were generated by top nodes (hub genes)- TMPRSS2 and ERG genes plus 16 genes located in their intergenic regions, and external linker nodes- the direct interactive proteins. Data on PPI were retrieved from 6 databases. Among them, 5 databases provided validated data on PPI: HPRD [16], DIP [17], IntAct [18], MINT [19,20], and APID [21], while one database provided predicted data on PPI: STRING [22]. Cytoscape_3.5.0 software [23] was utilized to plot the visualized diagram of PPI networks. We downloaded the data of PPI from multiple protein interaction databases mentioned above, and extracted the PPI information recorded more than twice, and formed the files with the extension CSV. Then, the files were imported into Cytoscape software to draw the PPI network maps.
The target genes of microRNAs located in the intergenic region of TMPRSS2 and ERG
microRNA sequences and their stem-loop structures were retrieved from the miRbase database [24,25]. microRNA locations were retrieved from UCSC Genome Browser on Human Assembly on Dec. 2013 (GRCh38/hg38), [26]. The target genes of intergenic microRNAs were predicted by four databases: TargetScanhuman7.1 [27] RNA22 [28,29], miRDB [30,31], and TargetMiner [32]. Cytoscape_3.5.0 software [23] was utilized to plot the visualized diagram of the networks between intergenic microRNAs and predicted target genes. In the same way, we extracted the information of the predicted target genes which were recorded more than twice, and formed the CSV files, and imported these files into Cytoscape software to draw the target gene network maps.
Gene ontology enrichment and signal pathway analysis
Gene annotation and gene ontology enrichment analysis of each nodal protein or each target protein were carried out via two databases: GO [33], and WEGO [34] (Web Gene Ontology Annotation Plot. The signaling pathways for TMPRSS2 and ERG and the intergenic genes were retrieved from six databases: KEGG [35], REACTOME [36,37], Wiki Pathways [38], PID [39,40] Biocarta [41], and Signal ink [42].
Transmission scanning electron microscope assay
We gained the human normal prostate epithelial cell line RWPE- 1 from American Type Culture Collection (ATCC). Cells were maintained in RPMI (Roswell Park Memorial Institute) medium (Sigma) supplemented with 10% FBS (fetal bovine serum). Then, RWPE-1 cells were transfected with Fugene 6 (Promega) using eukaryotic expression vector pcDNA3.1-T2E and control vector pcDNA3.1 (Promega). After cells steadily expressed TMPRSS2 - ERG fusion protein, we collected and fixed cells according to the experimental requirements and then cells were photographed using the transmission scanning electron microscopy.
Results
The difference between translocation-type and deletion-type lies in the fact that the latter is generated by the deletion of about 3.0 Mb region located between TMPRSS2 and ERG genes [43,44], and the role of these intergenic genes in PCa is not clear. When TMPRSS2 (chr 21:41,464,551-41,508,159) and ERG (chr 21: 38,380,027-38,498,483) were input into UCSC database [26] for query, 16 known genes and 3 microRNAs were mapped in the intergenic region. In the Supplementary Figure 1 and Supplementary Table 1, gene localizations and various splicing variants of the intergenic genes and micrRNAs were displayed, as well as the single nucleotide polymorphism (SNP) and conservation was shown.
microRNAs in the intergenic region
Supplementary Table 2 displayed the mature sequences and secondary stem-loop structures of microRNAs: miR-3197, miR- 4760, and miR-6508, which located in the intergenic region between TMPRSS2 and ERG genes.
Protein-protein interaction networks
According to the query in PPI databases, we sorted out 681 interactive proteins validated by experiments and 144 predicted proteins, which interacted directly with 18 proteins encoded by TMPRSS2 , ERG , and those intergenic genes. Among them, the validated PPI relations were retrieved from five databases: HPRD, DIP, IntAct, MINT and APID [16-21], while predicted PPI relations were obtained from the STRING database [22]. The PPI networks were built on the basis of the confirmed and predicted PPI data by Cytoscape3.5.0 software [23]. The PPI networks of validated proteins comprised of 1260 interactions from 699 proteins (network nodes), while the PPI networks of predicted proteins involved 1058 interactions from 162 protein nodes.
microRNAs in the intergenic regions and the target genes
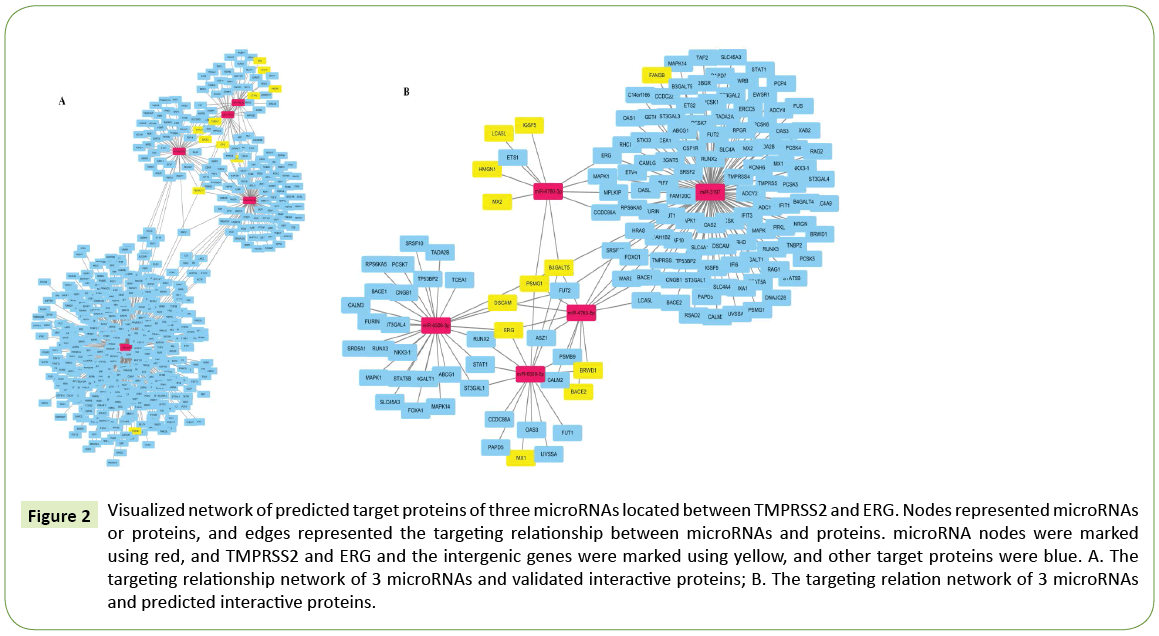
At present, there was no data on the target genes validated by experiments of microRNAs (miR-3197, miR-4760, and miR-6508) located in the intergenic region of TMPRSS2 and ERG . Data on the predicted target genes of three microRNAs were obtained from four databases: TargetScanhuman 7.1, RNA22, miRDB, and TargetMiner [27-32]. In combination with the above PPI network information, we used cytoscape_3.5.0 [23] software to draw the network diagrams of three microRNAs. The interactions of each nodal protein were derived from the verified PPI and the predicted PPI mentioned above, respectively, including 640 targeting relations with validated proteins and 179 targeting relations with predicted proteins. (Figure 2A) displayed the targeting relation networks of 3 microRNAs with the validated proteins. Among them, the relation network of miR-3197 was alienated from the other 2 microRNAs’. The relation networks of two splicing variants of miR-6508: miR-6508-5p and miR-6508- 3p were relatively independent from each other. Whereas two splicing variants of miR-4760 shared multiple common targets. Similar trend was reflected in the targeting relation networks of 3 microRNAs with the predicted proteins (Figure 2B). In addition to the above phenomena, the linkage between two splicing variants of miR-4760 was not apparent in (Figure 2B), which might be due to the limited data retrieval of the predicted target proteins.
Figure 2: Visualized network of predicted target proteins of three microRNAs located between TMPRSS2 and ERG. Nodes represented microRNAs or proteins, and edges represented the targeting relationship between microRNAs and proteins. microRNA nodes were marked using red, and TMPRSS2 and ERG and the intergenic genes were marked using yellow, and other target proteins were blue. A. The targeting relationship network of 3 microRNAs and validated interactive proteins; B. The targeting relation network of 3 microRNAs and predicted interactive proteins.
Analysis on gene ontology enrichment and signal pathways
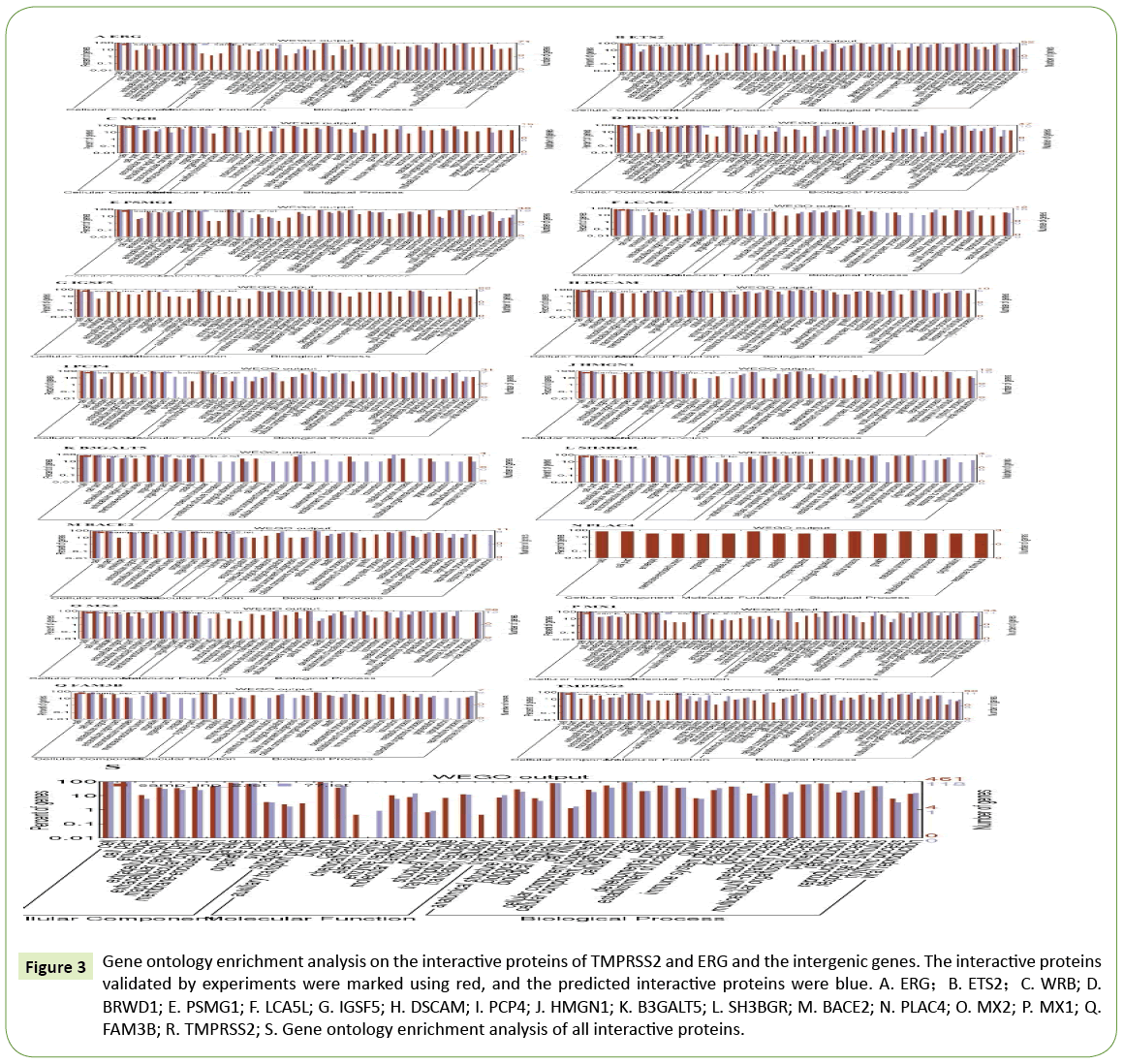
According to the findings from the visualized PPI networks, the PPI relations were relatively alienated from each other for the 18 genes, including TMPRSS2 and ERG , and 16 intergenic genes. These genes were further analyzed on gene ontology enrichment and signaling pathways. MX1 and MX2 proteins shared almost similar functions, mainly participating in the antiviral immunity (influenza, measles and hepatitis virus, etc). B3GALT5 protein was mainly involved in the biosynthesis of mucopolysaccharide and glycossphingolipid. HMGN1 protein was mainly involved in the DNA repair, including nucleotide excision repair and the formation of excision complex. The functions of ETS and DSCAM proteins were diversified, including the involvement in multiple signaling pathways (ERBB, MAPK, RAS, WNT, GnRH prolactin, etc.) and several types of cancers (colorectal, kidney and pancreas cancer, etc.) and a variety of infections (Shigella, Salmonella, pertussis, hepatitis and Toxoplasma gondii, etc.). SH3BR protein was involved not only in synaptic transmitter and morphine addiction, but also in the secretion of a variety of digestive fluids (saliva, gastric and pancreatic fluids, and bile). ERG and TMPRSS2 proteins were involved in cancer pathogenesis, and TMPRSS2 protein responded to multiple hormones (estrogen, glucocorticoids, and androgens), (Figure 3 and Supplementary Table 1).
Figure 3: Gene ontology enrichment analysis on the interactive proteins of TMPRSS2 and ERG and the intergenic genes. The interactive proteins validated by experiments were marked using red, and the predicted interactive proteins were blue. A. ERG;B. ETS2;C. WRB; D. BRWD1; E. PSMG1; F. LCA5L; G. IGSF5; H. DSCAM; I. PCP4; J. HMGN1; K. B3GALT5; L. SH3BGR; M. BACE2; N. PLAC4; O. MX2; P. MX1; Q. FAM3B; R. TMPRSS2; S. Gene ontology enrichment analysis of all interactive proteins.
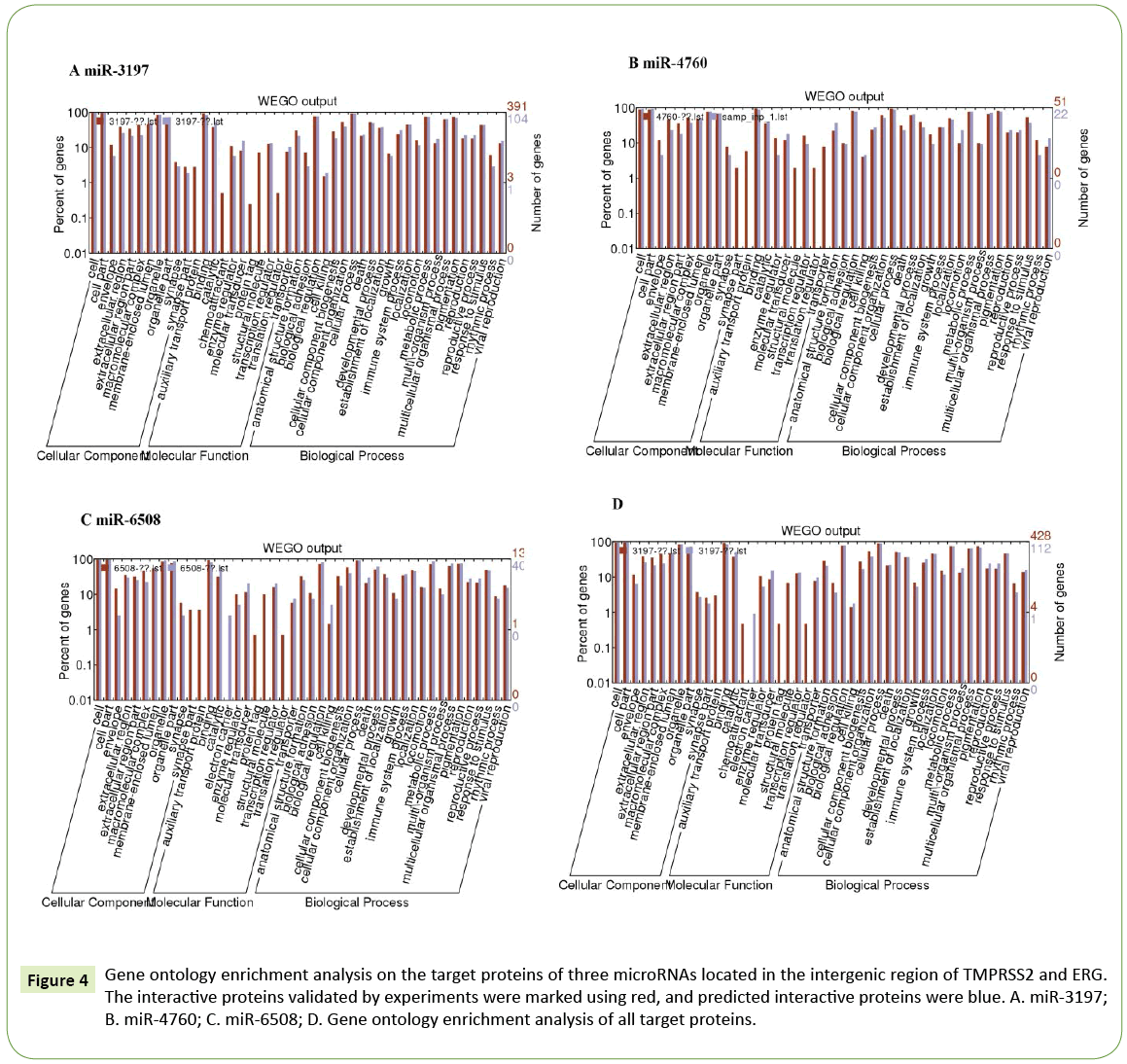
Gene ontology enrichment analysis was performed for the predicted target genes of three microRNAs (miR-3197, miR-4760, and miR-6508) located in the intergenic regions of TMPRSS2 and ERG . This revealed that the distribution and functions of three microRNAs were comprehensive and diversified. The three microRNAs showed no significant differences in cell distribution, function, and biological processes, except that miR-3197 was involved in chemotactic activity, while miR-6508 could be used as a protein tag (Figure 4).
Figure 4: Gene ontology enrichment analysis on the target proteins of three microRNAs located in the intergenic region of TMPRSS2 and ERG. The interactive proteins validated by experiments were marked using red, and predicted interactive proteins were blue. A. miR-3197; B. miR-4760; C. miR-6508; D. Gene ontology enrichment analysis of all target proteins.
Transmission scanning electron microscope assay
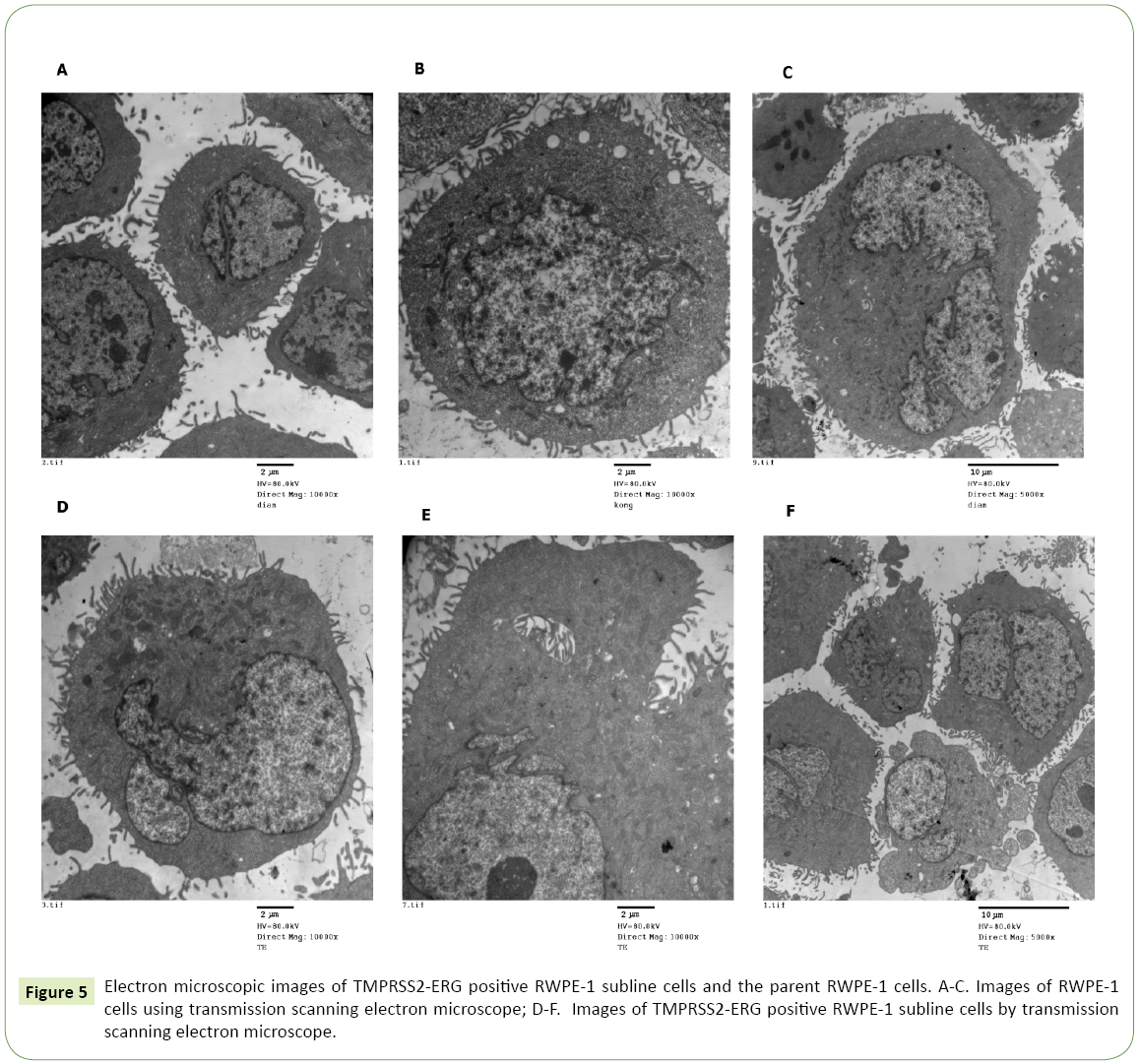
Finally, we compared the ultrastructure of TMPRSS2 -ERG positive RWPE-1 subline cells and the parent RWPE-1 line cells, and analyzed their differences. The parent RWPE-1 cells were closely arranged, small in size, with abundant surface microvilli and regular nuclear morphology. In cytoplasm, there were abundant mitochondria with clear crest membranes, abundant rough endoplasmic reticula, and a small amount of intracellular ton filaments (Figures 5A-5C). TMPRSS2 -ERG positive RWPE-1 cells were about twice as large as the parent cells with abundant microvilli and organelles, and a large amount of nuclear fission occurred (Figures 5D-5F). This finding was consistent with the complex multiple functions of TMRPSS2, ERG and the intergenic genes and microRNAs, which were involved in a wide range of cellular phenotypic regulation and biological processes.
Discussion
TMPRSS2-ERG gene fusion is specifically expressed in PCa, and deletion-type fusion causes the loss of the intergenic genes and the partial regions of TMPRSS2 and ERG genes. To describe the effects of these gene deletions in the progression of PCa, we obtained the functions of these genes from multiple bio information databases, as well as data on their direct interaction proteins. From the visualized PPI networks mapped using CytoScape software; these genes had their own interaction networks which were relatively independent of each other (Figure 1). The analyses of gene ontology enrichment and signal pathways demonstrated that other genes had no linkage with each other in functions and biological processes, except that MX1 and MX2 shared the similar functions, and ERG and TMPRSS2 were both involved in carcinogenesis. These genes were neither from the same family nor related genes with synergistic effects. We established TMPRSS2 -ERG positive RWPE-1 subline cells, and compared their ultrastructure with the parent RWPE-1 line cells by electron microscope. The fusion positive cells were two-fold larger than the parent cells, with abundant microvilli and organelles, and with increased nuclear fission. Additionally, the analyses of gene ontology enrichment and signal pathways implicated that these intergenic genes could play the critical roles in human cancers. In particular, ERG , ETS2 , BRWD1, DSCAM, HMGN1 and TMPRSS2 genes participated in the initiation and progression of a variety of cancers, including colorectal cancer, renal cell carcinoma, pancreatic cancer, endometrial cancer, glioma, PCa, thyroid cancer, bladder cancer, leukemia, melanoma and non-small cell lung cancer, etc. Among them, the deletion of HMGN1 led to severe DNA repair disability.
Linns team revealed the relationship between the intergenic genes (ETS2 and BACE2) with the recurrence and lethal phenotype of PCa. PTEN-/ETS2- double knock-out mice spontaneously developed prostate adenoma, suggesting that ETS2 was a tumor suppressor gene whose deletion deteriorated the disease [15]. The expression levels of ETS2 and BACE2 were significantly reduced in lethal PCa. Data from animal models and human PCa cell lines indicated that the deletion of ETS2 gene would result in PCa progression, combining with PTEN gene deletion.
Mx1, also known as MxA (Myxovirus resistance A), was another gene located in the intergenic region of TMPRSS2 and ERG . The gene encodes a kind of 78 kDa GTPase that is mediated by interferon (IFN). It was expressed in PC3 cells, but not in malignant metastatic PCa PC3M cells. Stable expression of exogenous Mx1 in PC3M cells could inhibit cell migration and invasion in vitro [45]. Meta-analysis suggested that the expression of Mx1 was inversely correlated with the progression of PCa. Compared with normal tissues, the expression of Mx1 was significantly reduced in PCa tissues. In agreement, Mx1 expression knock-down by shRNA in DU145 cells obviously promoted cell proliferation. Also, the overexpression of Mx1 in another PCa cell line, LNCaP, resulted in cell cycle arrest [46].
According to the previous literature, ETS2 and Mx1 were reported as relevant tumor suppressor genes in PCa, while HMGN1 gene was used in the diagnosis, prevention and treatment of PCa. High mobility group (HMG) is a nucleosome binding superfamily, including HMGA, HMGB, and HMGN subfamilies. And each subfamily contains several members, such as HMGB 1-3 and HMGN 1-5. HMGN1 (high-mobility group N1) is a member of HMGN subfamily and is commonly expressed in vertebral animal cells. HMGN1 is composed of two major domains: a C-terminal chromatin uncoiling domain, and an N-terminal nucleosome binding domain which specifically binds to 147 bp DNA sequences that winds to the histone core octamer [47]. It also modulates the chromatin structure. Over the past 35 years, HMGN1 was considered to bind to the transcriptional regulatory elements: the promoter and the enhancer [48]. The genome map of the embryonic fibroblasts of Hmgn1-/-, Hmgn2-/-, Hmgn1-/-n2-/- knockout mouse revealed that the deletion of HMGN variants led to the remodeling of the DNase I hypersensitive site. Therefore, we concluded that HMGN1 played an extremely critical role in ensuring the genetic markers of transcriptional regulatory regions, and transcription fidelity. In addition, HMGN1 had also an important role in repairing the damaged DNA [49].
In 2012, HMGN1 was recognized as a new alarmin [50]. Alarmin is an endogenous molecule that provides early alarm signals to the immune system to attract and activate leukocytes, in particular, the antigen presenting cells (APCs). HMGN1 promotes the maturation of dendritic cells and other APCs, ensuring preferential participation of antigen specific Th1 cells in the immune response [50]. In 2014, Wei and colleagues declared HMGN1 as a vaccine adjuvant that could help anti-tumor immune response [51]. Hmgn1tm1/tm1 mice, lacking the nucleosome binding domain of HMGN1, showed earlier onset of liver cancer phenotype than Hmgn1+/+ littermates after 2 weeks induction with nitrosodimethylamine injection [52]. Improper responses to the DNA in Hmgn1−/− cells or mice generated the accumulation of genome aberrations that were responsible for cancer progression [47]. A variety of proto-oncogenes, such as c-fos, Bcl-3, and N-cadherin, were upregulated in the case of Hmgn1−/− , while the expression of apoptotic gene NGFr was downregulated [53]. The number of spontaneous tumors was two-fold higher in Hmgn1−/− mice than in Hmgn1+/+ littermates. Primary cells purified from Hmgn1−/− mice proliferated faster than those from Hmgn1+/+ mice. In addition, when Hmgn1−/− tumor cells were inoculated into the nude mice, the tumor growth was 5-6 folds larger than the tumor cells with Hmgn1+/+ [54]. Hmgn1−/− mice were more prone to various malignant tumors and more likely to metastasize than Hmgn1+/+ mice [54]. In this regard, we concluded that HMGN1 modulates chromatin structure at the promoter and enhancer sites, thereby activating or inhibiting cancer-related genes: proto-oncogenes and tumor suppressor genes. At the same time, HMGN1 inhibited tumor growth via promoting DNA repair and genomic stability, and exerted the anti-tumor effects via stimulating anti-tumor immune response.
Our study was the first comprehensive functional analysis of TMPRSS2 and ERG genes and the intergenic genes and microRNAs from multiple bioinformatics platforms. We believed that similar strategies would be helpful to further elucidate complicated biological processes and had broad application prospects in cell biology and functional genomics. In summary, the recognized genes could benefit to understand the molecular mechanisms of carcinogenesis of TMPRSS2 -ERG positive PCa. In addition, further study of these genes could help identify the elevated risk of PCa due to TMPRSS2 -ERG gene fusion and deletion of the intergenic genes and microRNAs.
Conclusion
The study concerned the research hotspot in the field of PCa that aroused the attention of researchers since 2016. It was the first comprehensive functional analysis of the genes and microRNAs located between TMPRSS2 and ERG from multiple bioinformatics platforms. These genes had their own networks of interactions, and they were relatively independent of each other. The analyses of gene ontology enrichment and signal pathway demonstrated that there was no obvious connection between these genes. Otherwise, except that two splicing variants of miR-4760 shared a variety of common targets, the relationship networks of miR-3197 and miR-6508 were relatively independent from each other. This revealed that the distribution and functions of three microRNAs were comprehensive and diversified. Finally, the electron microscopy results showed that the cell volume of TMPRSS2 -ERG positive RWPE-1 cells was twice as large as that of the parent line cells, with abundant microvilli and organelles and nuclear fission. This phenomenon was consistent with the multifunction of TMRPSS2, ERG and the intergenic genes and microRNAs. These genes and microRNAs participated in a variety of cell phenotype regulation and biological processes.
To sum up, the study supposed that PCa might be drived by the fusion of TMPRSS2 and ERG genes, especially by the deletion of intergenic genes and microRNAs. According to the analysis results of this article, the functions of the intergenic genes and microRNAs were diversified and complex and they were mutually independent forming their respective PPI networks. The loss of their tumor suppressor roles might be the key reasons for the carcinogenesis of PCa.
Author's Contributions
RGM and GJJ participated in the download of the data from public databases. CH performed the statistical analysis and bioinformatics analysis. SCJ participated in the design of the study and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgement
The authors would like to acknowledge the professional writing services provided by MedSci Company. And thanks to Harbin medical university for transmission electron microscopy assay and morphological evaluation.
Availability of Data and Materials
All data generated or analysed during this study are included in this published article and its additional information files.
Funding
This work was supported by Zhejiang Provincial Science Technology Program of China (2013C33101), Zhejiang Medical Platform Program (2015RCA023), and Shaoxing municipal health and family planning science and technology innovation project (2017CX004).
References
- Siegel RL, Miller KD, Jemal A (2017) Cancer statistics. CA Cancer J Clin 67: 7-30.
- Qi JL, Wang LJ, Zhou MG, Liu YN, Liu JM, et al. (2016) Disease burden of prostate cancer among men in China, from 1990 to 2013. Zhonghua Liu Xing Bing Xue Za Zhi 37: 778-82.
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, et al. (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644-648.
- Kelly GM, Kong YH, Dobi A, Srivastava S, Sesterhenn IA, et al. (2015) ERG oncoprotein expression in prostate carcinoma patients of different ethnicities. Mol Clin Oncol 3: 23-30.
- Xu B, Chevarie-Davis M, Chevalier S, Scarlata E, Zeizafoun N, et al. (2014) The prognostic role of ERG immunopositivity in prostatic acinar adenocarcinoma: A study including 454 cases and review of the literature. Hum Pathol. 45: 488-497.
- Schaefer G, Mosquera JM, Ramoner R, Park K, Romanel A, et al. (2013) Distinct ERG rearrangement prevalence in prostate cancer: higher frequency in young age and in low PSA prostate cancer. Prostate Cancer Prostatic Dis 16: 132-138.
- Schneider TM, Osunkoya AO (2014) ERG expression in intraductal carcinoma of the prostate: comparison with adjacent invasive prostatic adenocarcinoma. Mod Pathol 27: 1174-1178.
- Tsourlakis MC, Stender A, Quaas A, Kluth M, Wittmer C, et al. (2016) Heterogeneity of ERG expression in prostate cancer: a large section mapping study of entire prostatectomy specimens from 125 patients. BMC Cancer 16: 641.
- Guo CC, Wang Y, Xiao L, Troncoso P, Czerniak BA (2012) The relationship of TMPRSS2-ERG gene fusion between primary and metastatic prostate cancers. Hum Pathol 43: 644-649.
- Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, et al. (2011) TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate 71: 489-497.
- Ren S, Peng Z, Mao JH, Yu Y, Yin C, et al. (2012) RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long non-coding RNAs and aberrant alternative splicings. Cell Res 22: 806-821.
- Jiang H, Mao X, Huang X, Zhao J, Wang L, et al. (2016) TMPRSS2: ERG fusion gene occurs less frequently in Chinese patients with prostate cancer. Tumour Biol 37: 12397-12402.
- Mao X, Yu Y, Boyd LK, Ren G, Lin D, et al. (2010) Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res 70: 5207-5212.
- Dong J, Xiao L, Sheng L, Xu J, Sun ZQ (2014) TMPRSS2: ETS fusions and clinic pathologic characteristics of prostate cancer patients from Eastern China. Asian Pac J Cancer Prev 15: 3099-3103.
- Linn DE, Penney KL, Bronson RT, Mucci LA, Li Z (2016) Deletion of Interstitial Genes between TMPRSS2 and ERG Promotes Prostate Cancer Progression. Cancer Res 76: 1869-1881.
- https://www.hprd.org/.
- https://dip.mbi.ucla.edu/dip/.
- https://www.ebi.ac.uk/intact.
- https://mint.bio.uniroma2.it/mint/.
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, et al. (2012) MINT, the molecular interaction database: update. Nucleic Acids Res 40: 857-861.
- https://cicblade.dep.usal.es:8080/APID/ init.action.
- https://string-db.org/.
- https://www.cytoscape.org/.
- https://www.mirbase.org/.
- Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: 68-73.
- https://genome.ucsc.edu/.
- https://www.targetscan.org/ vert_71/version7.1.
- https://cm.jefferson.edu/rna22/.
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, et al. (2006) A pattern-based method for the identification of MicroRNAbinding sites and their corresponding heteroduplexes. Cell 126: 1203-1217.
- https://mirdb. org/.
- Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43: 146-152.
- https://www.isical.ac.in/~bioinfo_miu/targetminer20.htm.
- 33 https://geneontology.org/.
- https://wego.genomics.org.cn/.
- https://www.kegg.jp/.
- https://www.reactome.org.
- Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, et al. (2011) Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 39: 691-697.
- https://www.wikipathways.org/index.php/WikiPathways.
- https://pid.nci.nih.gov.
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, et al. (2009) PID: the Pathway Interaction Database. Nucleic Acids Res 37: 674-679.
- https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways.
- https://www.signalink.net/.
- Burdova A, Bouchal J, Tavandzis S, Kolar Z (2014) TMPRSS2-ERG gene fusion in prostate cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 158: 502-510.
- Iljin K, Wolf M, Edgren H, Gupta S, Kilpinen S, et al. (2006) TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res 66: 10242-10246.
- Mushinski JF, Nguyen P, Stevens LM, Khanna C, Lee S, et al. (2009) Inhibition of tumor cell motility by the interferon-inducible GTPase MxA. J Biol Chem 284: 15206-15214.
- Brown SG, Knowell AE, Hunt A, Patel D, Bhosle S, et al. (2015) Interferon inducible antiviral MxA is inversely associated with prostate cancer and regulates cell cycle, invasion and docetaxel induced apoptosis. Prostate 75: 266-279.
- Gerlitz G (2010) HMGNs, DNA repair and cancer. Biochim Biophys Acta 1799: 80-85.
- Martínez de Paz A, Ausió J (2016) HMGNs: The enhancer charmers. Bioessays 38: 226-231.
- Deng T, Zhu ZI, Zhang S Postnikov Y, Huang D, et al. (2015) Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers. Genome Res 25:1295-1308.
- Yang D, Postnikov YV, Li Y, Tewary P, De la Rosa G, et al. (2012) High-mobility group nucleosome-binding protein 1 acts as an alarmin and is critical for lipopolysaccharide-induced immune responses. J Exp Med 209: 157-171.
- Wei F, Yang D, Tewary P, Li Y, Li S, et al. (2014) The Alarmin HMGN1 contributes to antitumor immunity and is a potent immunoadjuvant. Cancer Res 74: 5989-5998.
- Postnikov YV, Furusawa T, Haines DC, Factor VM, Bustin M (2014) Loss of the nucleosome-binding protein HMGN1 affects the rate of N-nitrosodiethylamine-induced hepatocarcinogenesis in mice. Mol Cancer Res 12: 82-90.
- Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, et al. (2004) Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell 15: 573-584.
- Birger Y, Catez F, Furusawa T, Lim JH, Prymakowska-Bosak, et al. (2005) Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res 65: 6711-6778.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences