BI(G)MED as a Very Novel Method Based on Oral Immunogenetic Therapy to Prevent and Treat Allergic Diseases Especially Asthma
Gilbert Glady*
Department of Internal Medicine, European Bio Immune(G)ene Medecine Association, France
- *Corresponding Author:
- Gilbert Glady
Department of Internal Medicine
European Bio Immune(G)ene Medecine Association, France.
Tel: +33 635 56 21 48
E-mail: info@ebma-europe.com
Received date: October 04, 2017; Accepted date: November 14, 2017; Published date: November 20, 2017
Citation: Allergic asthma; Innovative sublingual immunotherapy; microRNAs; Systems biology; Th1/Th2 balance
Abstract
Background: Oral immunotherapy has become a recognized method, validated by numerous publications and its use has developed in the current practice of an allergy clinic. We propose an immunotherapy administered by the sublingual route which is both more flexible than the immunotherapies already known for systemic use, devoid of any risk of side effects and equally effective in terms of therapeutic efficacy. The aim of such a therapy will clearly be that of a slow regulation of the molecular imbalances in relation to the state of allergy and a progressive recovery of the cellular homeostasis.
Methods and findings: For this purpose, we first resort to a regulation taking place at the epigenetic level thanks to the introduction in our therapeutic methodology of microRNAs used for their remarkable posttranscriptional regulatory action. All components of our cures are prepared in ultra-low doses to avoid any noxious side effects and administered per sublingual way. But the BioImmune(G)ene Medicine - BI(G)MED in shortcut - is also holistic in the sense that it does not just act in a regulating way on the immunogenetic mechanisms involved directly in the genesis of allergic diseases, but also search for neutralizing infectious, viral and bacterial and even fungal agents, which have a pernicious influence on the course of clinical allergic phenomena. To achieve this result, the BI(G)MED uses a wide range of biological tests to measure the patient's reaction potential at the immunogenetic level and the impact of infectious agents in direct interaction with the immune system.
Conclusion: The description of clinical cases with their therapeutic modalities will make it possible to highlight the regulatory properties of this new method referring to the self-regulation potential inherent in cellular homeostasis. Keywords: Allergic asthma; Innovative sublingual immunotherapy; microRNAs; Systems biology; Th1/Th2 balance
Keywords
Allergic asthma; Innovative sublingual immunotherapy; microRNAs; Systems biology; Th1/Th2 balance
Introduction
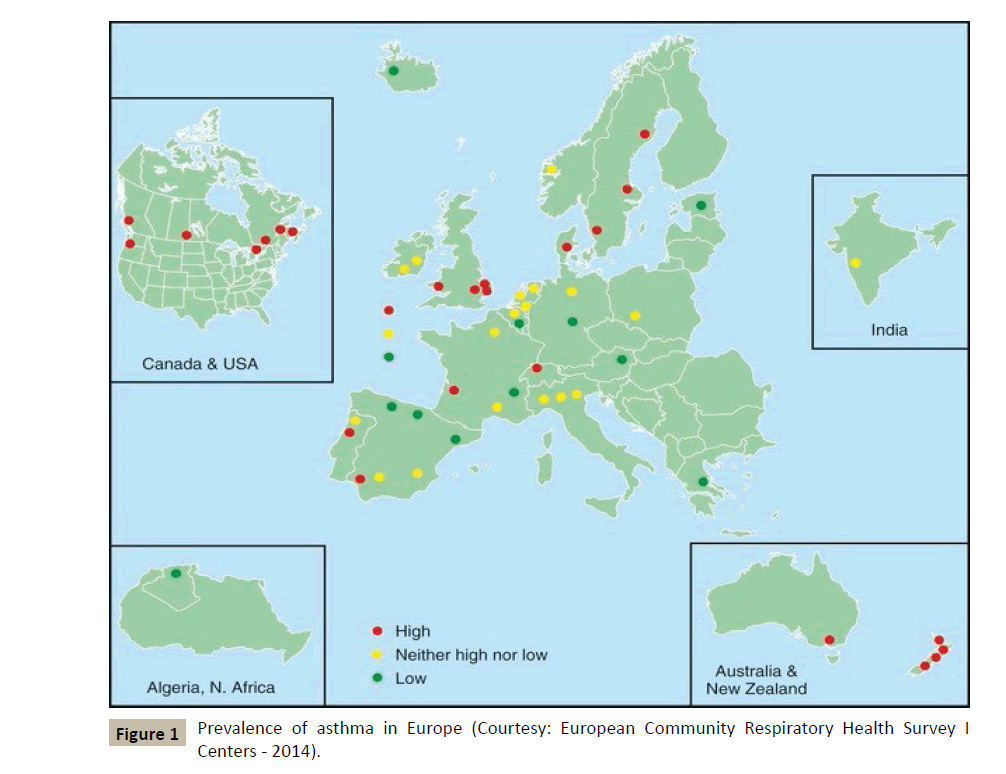
According to the WHO, nearly 300 million people suffer from asthma in the world, and the incidence of this disease and allergic conditions in general is increasing in the so-called industrialized countries (Figure 1). Asthma is thus becoming a growing problem in the field of public health, if only because of the increasing costs it generates in social communities.
Figure 1 :Prevalence of asthma in Europe (Courtesy: European Community Respiratory Health Survey I Centers - 2014).
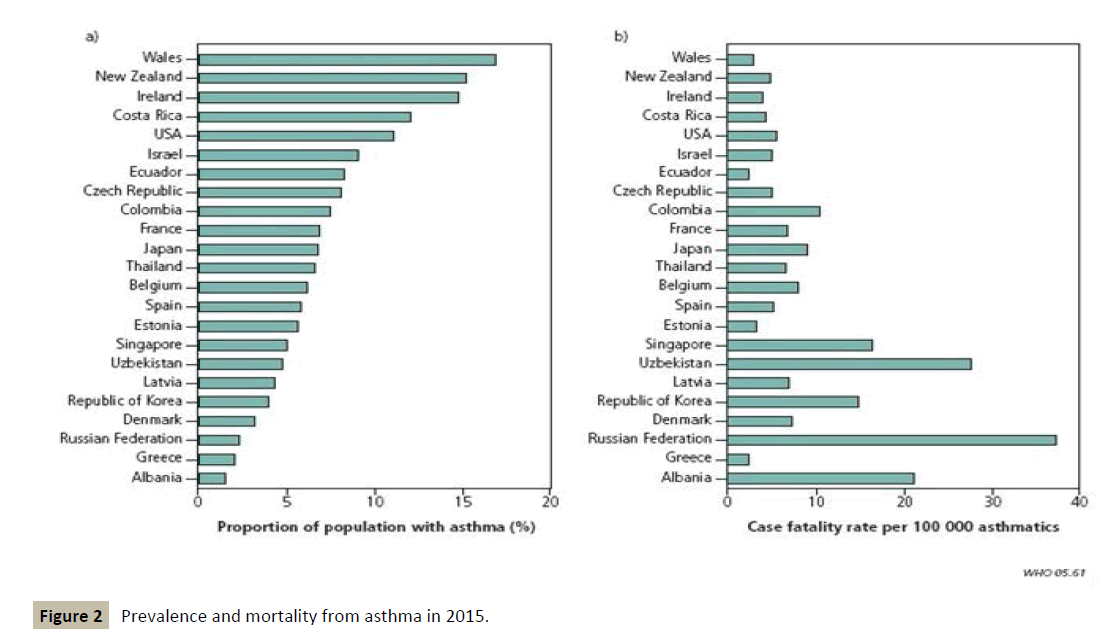
Moreover, the prognosis of the disease remains worrying in the medium to long term, suggesting that at present there is no treatment capable of slowing down the progression or even stopping its evolution (Figure 2).
Figure 2 :Prevalence and mortality from asthma in 2015.
One reason among others may be the absence of therapeutic methods taking into account the totality of the pathophysiological factors involved in the development of this bronchopulmonary disease in order to regulate them concomitantly.
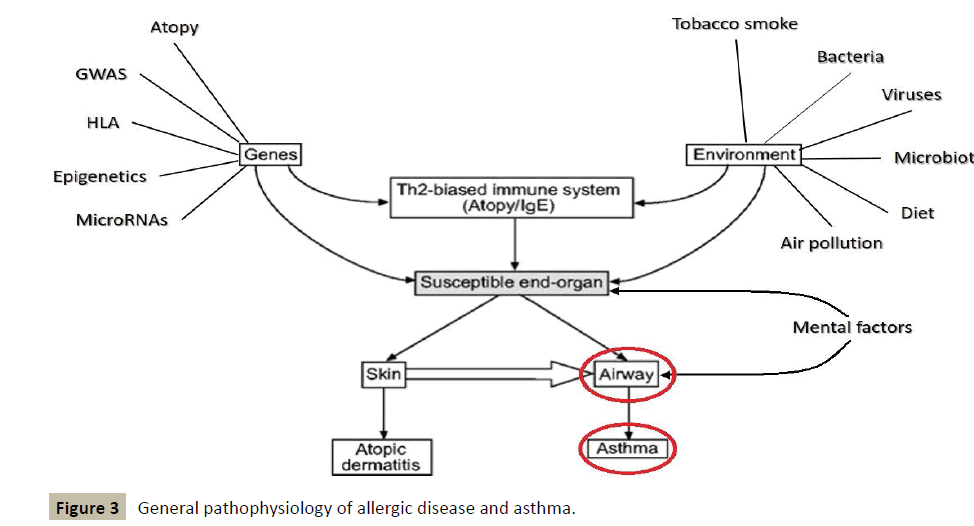
The following figure, which shows the general pathophysiology of allergic disease and asthma, somehow traces the contours of a diagnostic and therapeutic approach of holistic type (Figure 3).
It places a particular emphasis on the conditions necessary for a regulatory treatment, whose impact must take place at the level of genes and consider environmental factors, before seeking to regulate the homeostatic immune balance TH1/TH2.
Generally in allergy, the first stage of sensitization takes place at the level of the intestinal barrier following a dysfunction or a mechanism of rupture of genetic and/or environmental origin affecting the latter. This impairs an alteration of the integrity of this intestinal barrier, which will promote sensitization to very common antigens then becoming allergens.
At the level of cellular biology, all these processes will facilitate the implementation of predominantly TH2 reactions associated with high levels of IgE, activation of the main cell types involved in allergy and the establishment of a corresponding phenotype in the major organs of the allergic diathesis.
To try to achieve this objective, there is at present a very fruitful concept, that of "systems biology" and its complementary “P4 medicine” developed by Hood and Tian from the Institute for Systems Biology (ISB) in Seattle, which coined the term “P4 medicine” (Personalized, Predictive, Preventive and Participatory medicine) some years ago to reach a global approach of biological phenomena and a holistic approach involving the patient in his organo-psycho-social totality as well [1].
The ISB studies biological complexity based on three fundamental premises [2]: (i) there are two types of biological information, i.e., digital genome information and environmental information, outside the genome, that modifies the above-mentioned digital information; (ii) biological information is captured, processed, integrated and transferred by means of biological networks (RNA, proteins, controlling regions of the genes and small molecules) to the molecular systems that execute vital functions; and (iii) biological information is codified in a multi-scale hierarchy: DNA, RNA, proteins, interactions, biological networks, tissues and organs, individuals and, finally, ecologies. It is important to highlight the role that environmental factors can play as regulators that can interfere at all levels of involvement in this broad network of information coming from the genome.
Systems biology can be defined as a modelling of complex biological processes on a computational and mathematical database. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach (holism instead of the more traditional reductionism) to biological research.
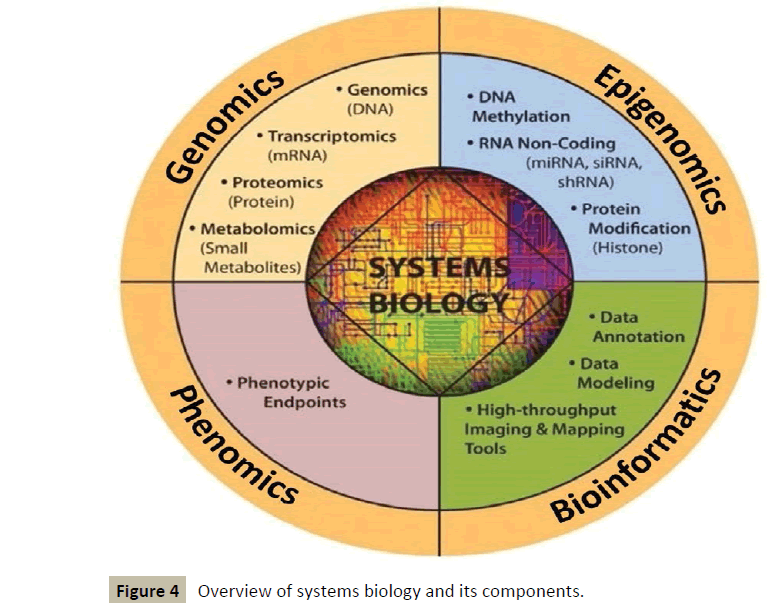
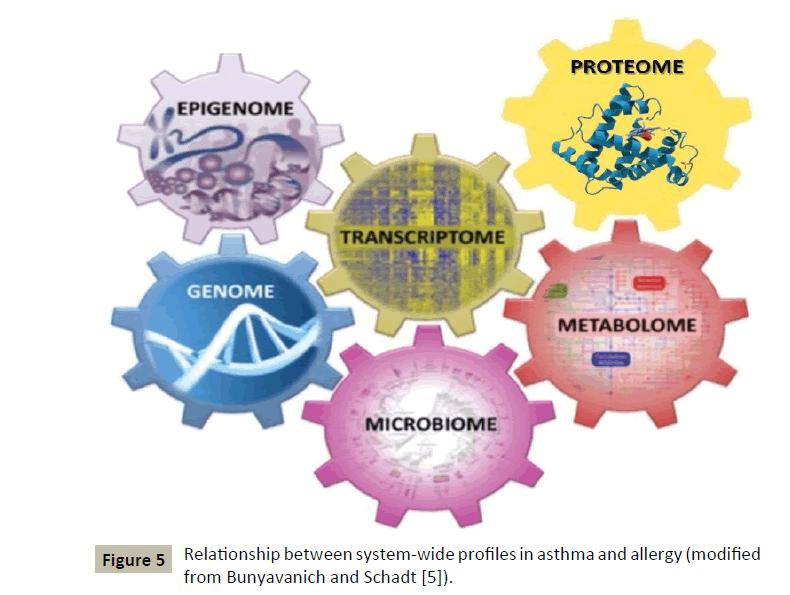
It works in an interdisciplinary biology-based research area focusing on the complex interactions between biological systems of living organisms. It uses a holistic approach replacing a traditionally reductionist approach, allowing it to take into account all the molecular components of the cell (nucleic acids, proteins...) and their pathophysiologic interactions as well [3]. This new way of approaching the core of diseases will make use of various tools [4], which will include a lot of “omics” such as genomics, epigenomics, proteomics, metabolomics, etc. (Figure 4).
Figure 4 :Overview of systems biology and its components.
Moreover, this concept of "systems biology" corresponds entirely to the approach to human diseases that has been used for many years in the framework of BI(G)MED, and that we are able to translate at both the diagnostic and therapeutic level.
This innovative concept, although identified in another context by some authors a long time ago, has been applied to allergic diseases in general and more particularly to asthma [5] (Figure 5) by two American authors whose demonstration will be followed step by step:
Figure 5: Relationship between system-wide profiles in asthma and allergy (modified from Bunyavanich and Schadt [5]).
Asthma & the genome
GWAS (Genome-Wide Association Study) identified the 17q21 locus as the one associated with asthma with the greatest reproducibility. This locus contains four genes (abbreviated as ORMDL3, GSDMB, ZPBP2 and IKZF3) which will act as endogenous inducers of inflammation via their effects on the endoplasmic reticulum [6-9].
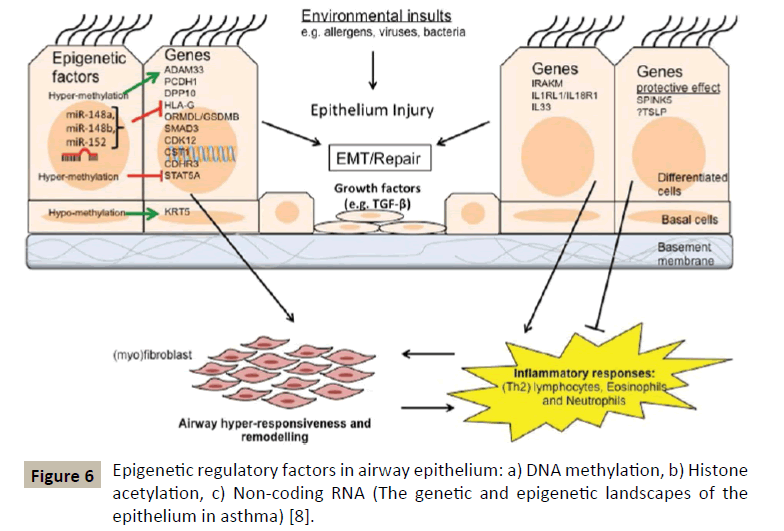
GWAS has been able to identify other asthma susceptibility loci (Figure 6) among which we will find genes involved in the recruitment and/or activation of pro-inflammatory cells (TSLP, IL33, IL1RL1), T-cell responses and differentiation (HLA, IL2RB, DENND1B, IL6R) and modulation of certain cell signalling pathways (PDE4D, SMAD3, CDHR3).
But GWAS has by no means made it possible to identify exhaustively the risk of asthma; other methods such as positional cloning in combination with linkage analysis have allowed the detection of other genes associated with the development of asthmatic disease in the airways [10,11]. They included ADAM33, NPSR1 also called GPRA, PCDH1, SPINK5, IRAKM, DPP10 and HLA-G genes.
That’s how, first of all, ADMA33 localized on the 20p13 locus [12], was found to be involved in the remodelling of the bronchial musculature and hyper responsiveness was confirmed.
Asthma & the transcriptome
In general, transcriptomics is a more dynamic approach to pathophysiological processes taking place in tissues than GWAS, whose approach it completes synergistically.
To distinguish healthy from asthmatic subjects, RNA-sequencing techniques can be used on tissue samples from the lower and upper airways. Using these techniques on bronchial biopsies a Dutch team was able to demonstrate a significantly different expression between asthma and controls for 46 genes including those of pendrin, periostin and Bcl-2 [13].
A transcriptomic profile of asthma was also obtained from peripheral blood samples and demonstrated in patients with allergic asthma an increased IL4R expression in circulating CD19+ B-lymphocytes compared to controls, suggesting that a possible transcriptional dysregulation of B-lymphocytes is implicated in allergic asthma.
Using a polyA-selected whole transcriptome sequencing technique, a US team has very recently been able to identify a very large population of children free from any respiratory disease, carrying pathogenic and transcriptionally active respiratory viruses [14].
Elinav and his team have recently demonstrated that by combining sRNA expression profiles from the oral microbiome with mRNA expression data, it was shown that sRNAs are potentially involved in the regulation of metabolic activities of the oral microbiome and, in this way, in the evolution of a commensal oral microbiome to a dysbiotic microbiome [15].
Asthma & the proteome
Proteomics analyses the structure and function of biological systems. Proteins provide intricate control of cellular machinery, and are in many cases components of that same machinery; they can thus represent excellent biomarkers. A proteome is a set of proteins produced in an organism, system or biological context. The proteome is not constant; it differs from cell to cell and changes over time. To some degree, the proteome reflects the underlying transcriptome.
Recently, various inflammatory biomarkers of airways obtained in a non-invasive manner in the exhaled air, blood and urine, have been identified as useful tools for the diagnosis and management of asthmatic disease [16,17] (Table 1).
| Exhaled breath | Exhaled breath condensate | Induced sputum | Blood |
|---|---|---|---|
| NOS2 | H2O2, LTB4, Cys-LT, ET-1, CCL11, MMP-9 | ECPs, IL-4, IL-5, IL-13, TNF-a, IL-6, GM-CSF, IL-12 | IL-3, IL-18, FGF, HGF, SCGF-ß, periostin |
Table 1 Airway inflammatory biomarkers in different sources.
All these molecules are detected and increased in asthma patients. Of course, all these inflammatory molecules will be able to be integrated with the appropriate dilutions in the BI(G)MEDformulas aimed at treating asthma.
Asthma & the epigenome
For the study of a disease it is necessary to complete the genomic and transcriptomic approaches by studying the epigenetic changes, which may characterize alterations that are independent from the DNA sequence and which may cause transcriptomic variations and modulate a downstream phenotype [18].
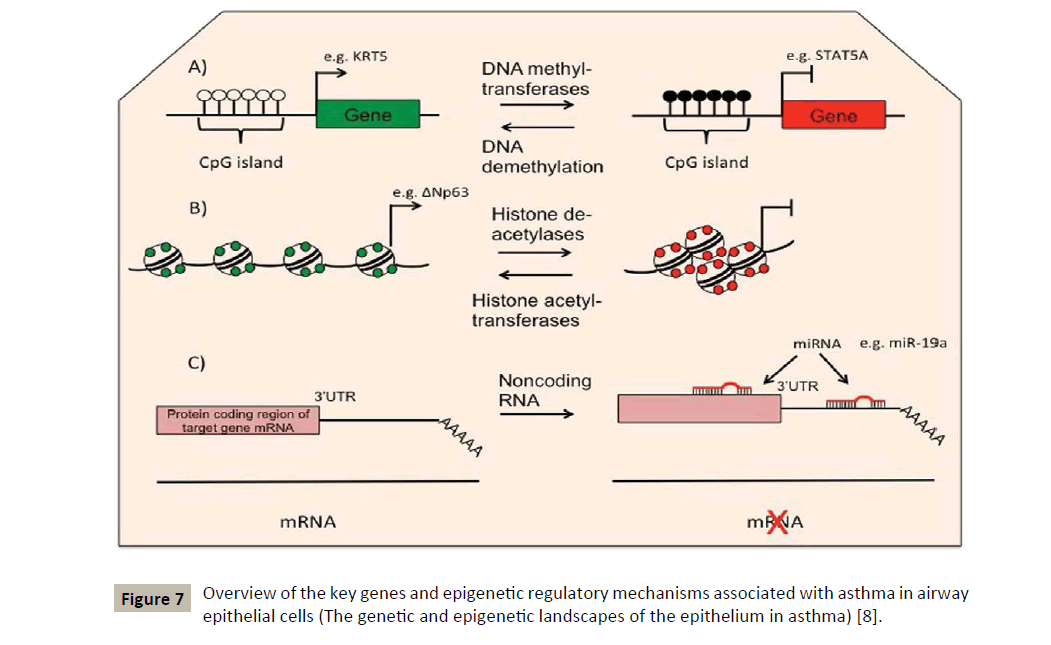
Usually epigenetic changes include processes of DNA methylation and hydroxymethylation, histone modifications and RNA interference by microRNAs [7] (Figure 7). The latter being extremely important in regulating the TH1/TH2 balance and in regulating allergic inflammation as well [19]. The interest shown for the observable epigenetic changes in allergic diseases and asthma comes from the fact that these modifications can serve as mediators to link exposure to certain epidemiologically identified environmental factors and the phenotype of asthmatic disease.
For our part, we pay particular attention to the RNA-interference as the fastest and perhaps best performing epigenetic process. It is also the easiest to implement for therapeutic purposes.
Asthma & the microbiome
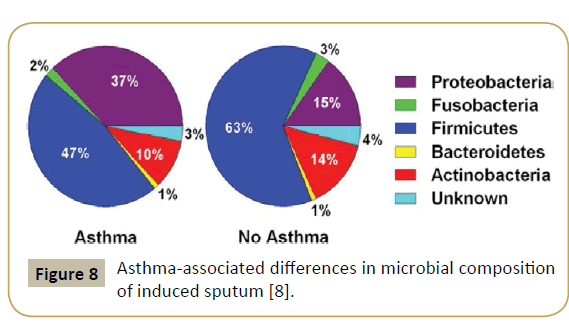
An approach using systems biology highlighted the role played by the microbiome and its interaction with the genome in asthma and allergy, showing in particular that the bronchial tree was physiologically not sterile. Thus, the latter contains in average the genomes of 2,000 bacteria/cm2 with a predominance of Firmicutes [20]. On the other hand, asthmatic subjects present a higher number of Proteobacteria (Figure 8). In 2015, a US team could demonstrate that specific microbiota is associated with, and may modulate inflammatory processes in patients with severe asthma and related phenotypes [21]. Airway dysbiosis in patients with severe asthma appears to differ from that observed in those with milder asthma in the setting of inhaled corticosteroid use.
In contrast to the great diversity in airway microbiota observed in asthma, a low diversity of intestinal microbiota in childhood may have been correlated with asthma occurrence in children at 7 years of age. Data from Canadian Healthy Infant Longitudinal Development (CHILD) Study (n=319) demonstrated that gut microbial dysbiosis during the first 3 months of life predisposed infants to a relatively high risk of asthma, which was associated with significant decreases in bacterial genera Faecalibacterium, Lachnospira, Veillonella and Rothia as well as reduced faecal acetate levels.
Asthma & the metabolome
The aim of metabolomic techniques is to collect low molecular weight compounds in biological samples, and to systematically evaluate, identify and quantify them using pattern recognition methods.
Recent studies have been conducted in a metabolomic context in the framework of asthma using condensed exhaled air, urine, serum and plasma [22]. Through these studies, different metabolites from several molecular pathways could be used to allow discrimination between asthmatics and control subjects, between asthma in crisis and stable asthma and finally between severe asthma or not.
A work published in 2017 concluded that « asthma is characterised by a modest systemic metabolic shift in a disease severitydependent manner and that steroid treatment significantly affects metabolism” [23].
Interesting is also a study of another group, published very recently in 2017, which emphasizes the importance of the relationships between inflammatory mechanisms in the airways and ROS (Reactive Oxygen Species) from a therapeutic perspective [24].
Materials and Methods
The BI(G)MED method is now ten years old, it is not yet the age of reason, but we approach it very slowly. It expanded at the same time as the systems biology, based on the same holistic principles on one side in its diagnostic approach where we use, e.g. a lymphocyte typing to get an overview of the cellular immunity and a protein profile to appreciate humoral immunity. In this way, it combines the most accurate detection of the immune state in relation to a lot of environmental factors (microbial and allergic in particular). On the other side from a therapeutic point of view, it uses a very similar overall approach enabling it to regulate a vast molecular network disturbed in its functioning at the different levels mentioned above.
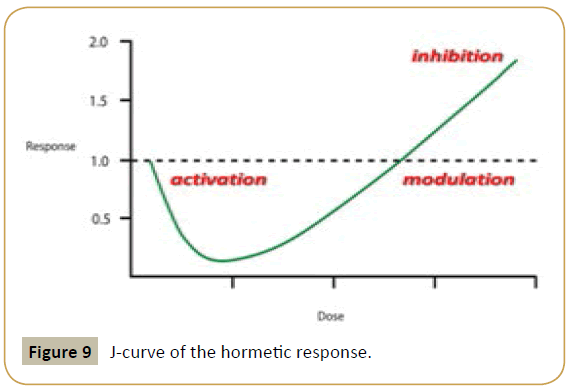
This is made possible by the use of ultra-low doses of all the molecular components involved. According to the general principle of Hormesis [25] and its corollary - the law of inversion of action according to dilution [26,27] – we will be able to direct the reaction in one or another orientation (i.e., activation, modulation or inhibition) (Figure 9).
Several types of studies have elucidated the various signalling pathways and molecular mechanisms involved in the phenomenon of Hormesis, which will generally involve enzymes such as kinases and deacetylases, as well as transcriptional factors such as Nrf-2 and NF-kappaB. The result will be an increased production of proteins capable of protecting and restoring the cells including growth factors, phase 2 and antioxidant enzymes, as well as chaperone proteins.
To be sure that all the molecular signals are adequately transmitted, we have selected xylitol nano vectors used sublingually (Figure 10) and able to transmit their message to the immuno-competent cells and molecules present in the oropharyngeal mucosa. We have thus developed three different therapeutic formulas, so-called Bioimmun(G)ene Regulators (Bimuregs), targeting both the TH1/TH2 balance, the regulation of eosinophils, and all the molecular networks involved at each level in the onset and progression of asthmatic disease (Table 2).
| TH2-REG | Molecule | Concentration | ||
|---|---|---|---|---|
| TNF-a | 1 × 10-10 mol | |||
| miR-126 | 2 | |||
| miR-375 | 2 | |||
| IL-4 | 2 | |||
| IL-5 | 2 | |||
| IL-13 | 2 | |||
| IL-25 (IL-17E) | 2 | |||
| IL-33 | 2 | |||
| TLR4 | 2 | |||
| ICOS/B7RP1 | 2 | |||
| B7-2/CD28 | 2 | |||
| OX40/OX40-L | 1 × 10-8 mol | |||
| CTLA-4 promoter & gene | 2 | |||
| TGF-ß | 1 × 10-6 mol | |||
| miR-21 | 2 | |||
| IL-10 | 1 × 10-8 mol | |||
| TSLP | 1 × 10-10 mol | |||
| STAT6 | 2 | |||
| RESPIREG–C1 | Molecule | Concentration | ||
| ADAM 33 (gene) | 1 × 10-8 mol | |||
| ERK 1-2 MAPK | 1 × 10-10 mol | |||
| PI3K | 2 | |||
| ITK | 2 | |||
| NGF | 2 | |||
| LTC4 | 2 | |||
| ADAM 8 (protein) | 2 | |||
| ß-arrestin-2 | 2 | |||
| TLR4 | 2 | |||
| TIMP-1 | 1 × 10-6 mol | |||
| MMP-9 | 1 × 10-10 mol | |||
| CCL11/CC24/CCL26/CCR3 | 2 | |||
| miR-16 | 2 | |||
| miR-26a | 2 | |||
| miR-106a | 2 | |||
| miR-126 | 2 | |||
| miR-140 | 2 | |||
| miR-145 | 2 | |||
| miR-146a/b | 2 | |||
| miR-181a | 2 | |||
| let-7 | 1 × 10-6 mol | |||
| miR-25 | 2 | |||
| miR-28-5p | 2 | |||
| miR-126 | 2 | |||
| miR-133a | 2 | |||
| miR-143 | 2 | |||
| miR-672 | 2 | |||
| GUT REG/MIR | Molecule | Concentration | ||
| IL-6 | 1 × 10-10 mol | |||
| IL-8 | 2 | |||
| IL-21 | 2 | |||
| IL-31 | 2 | |||
| IL-13 | 1 × 10-8 | |||
| miR-19b | 1 × 10-10 mol | |||
| miR-505 | 2 | |||
| miR-629 | 2 | |||
| miR-16 | 1 × 10-10 mol | |||
| miR-20a | 2 | |||
| miR-23b | 2 | |||
| miR-106 | 2 | |||
| miR-191 | 2 | |||
| miR-199a-5p | 2 | |||
| miR-223 | 2 | |||
| miR-340 | 2 | |||
| miR-362-3p | 2 | |||
| miR-532-3p | 2 | |||
| miR-594 | 2 | |||
| miRplus-1271 | 2 | |||
Table 2 Bimuregs used in the treatment of asthmatic disease.
Discussion
Asthma is a complex disease requiring a multidisciplinary approach as well as global therapeutics. Systems biology, and its extension P4 medicine, are very modern concepts that apply perfectly to the diagnosis and treatment of asthma and allergy in general.
The BI(G)MED method represents a practical application of these two concepts. A regulatory treatment is made possible by the use of a large number of immunocompetent molecules at ultra-low doses that can be synthesized with nanobiotechnology-derived techniques. These ultra-low doses of molecules are capable, according to the hormesis rules, to have an effective regulatory effect and protective role on the cellular structures involved in asthmatic disease. It is therefore possible to treat a chronic disease such as asthma over the long term without having to fear adverse effects.
It would be desirable and of great interest for the next few years to see the development of rigorous fundamental research in the field of ultramolecular medicine, because it seems to be a very promising challenge, which the use of known techniques in nanomedicine should make easily accessible.
References
- Hood L, Tian Q (2012) Systems approaches to biology and disease enable translational systems medicine. Genomics Proteomics Bioinformatics 10: 181-185.
- Bengoechea JA (2012) Infection systems biology: From reactive to proactive (P4) medicine. Int Microbiol 15: 55-60.
- Wang K, Lee I, Carlson G, Hood L, Galas D (2010) Systems biology and the discovery of diagnostic biomarkers. Dis Markers 28: 199-207.
- Zhang L, McHale CM, Rothman N, Li G, Ji Z, et al. (2010) Systems biology of human benzene exposure. Chem Biol Interact 184: 86-93.
- Bunyavanich S, Schadt EE (2015) Systems biology of asthma and allergic diseases: A multiscale approach. J Allergy Clin Immunol 135: 31-42.
- Portelli MA, Hodge E, Sayers I (2015) Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin Exp Allergy 45: 21-31.
- Akhabir L, Sandford AJ (2011) Genome-wide association studies for discovery of genes involved in asthma. Respirology 16: 396-406.
- Moheimani F, Hsu AC, Reid AT, Williams T, Kicic A, et al. (2016) The genetic and epigenetic landscapes of the epithelium in asthma. Respir Res 17: 119.
- Tamari M, Tanaka S, Hirota T (2011) Genome-wide association studies of asthma. Allergol Int 60: 247-252.
- March ME, Sleiman PM, Hakonarson H (2011) The genetics of asthma and allergic disorders. Discov Med 11: 35-45.
- Ober C, Yao TC (2011) The genetics of asthma and allergic disease: A 21st century perspective. Immunol Rev 242: 10-30.
- Tripathi P, Awasthi S, Gao P (2014) ADAM metallopeptidase domain 33 (ADAM33): A promising target for asthma. Mediators Inflamm 572025.
- Yick CY, Zwinderman AH, Kunst PW, Grünberg K, Mauad T, et al. (2013) Transcriptome sequencing (RNA-Seq) of human endobronchial biopsies: Asthma versus controls. Eur Respir J 42: 662-670.
- Wesolowska AA, Everman JL, Davidson R, Rios C, Herrin R, et al. (2017 Jan 19) Dual RNA-seq reveals viral infections in asthmatic children without respiratory illness which are associated with changes in the airway transcriptome. Genome Biol 18: 12.
- Bashiardes S, Zilberman-Schapira G, Elinav E (2016) Use of metatranscriptomics in microbiome research. Bioinform Biol Insights 10: 19-25.
- Paone G, Leone V, Conti V, De Marchis L, Ialleni E, et al. (2016) Blood and sputum biomarkers in COPD and asthma: A review. Eur Rev Med Pharmacol Sci 20: 698-708.
- Terracciano R, Pelaia G, Preianò M, Savino R (2015) Asthma and COPD proteomics: current approaches and future directions. Proteomics Clin Appl 9: 203-220.
- Bégin N (2014) Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin Immunol 10: 27.
- Pua HP, Ansel KM (2015) MicroRNA regulation of allergic inflammation and asthma. Curr Opin Immunol 36: 101-108.
- Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD (2013) Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol 131: 346-352.
- Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, et al. (2015) The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol 136: 874-884.
- Muñoz X, Bustamante V, Lopez-Campos JL, Cruz MJ, Barreiro E (2015) Usefulness of noninvasive methods for the study of bronchial inflammation in the control of patients with asthma. Int Arch Allergy Immunol 166: 1-12.
- Reinke SN, Gallart-Ayala H, Gómez C, Checa A, Fauland A, et al. (2017) Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J 49: 1601740.
- Qu J, Li Y, Zhong W, Gao P, Hu C (2017) Recent developments in the role of reactive oxygen species in allergic asthma. J Thorac Dis 9: E32-E43.
- Mattson MP (2008) Hormesis defined. Ageing Res Rev 7: 1-7.
- Calabrese EJ, Mattson MP (2017) How does hormesis impact biology, toxicology and medicine? NPJ Aging Mech Dis 3: 13.
- Al'mina NP, Mal'tseva EL, Chasovskaia TE (2014) Effect of dilute solutions of biologically active substances on cell membranes. Biofizika 59: 704-716.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences