Assessment of Molecular Diversity among Maize Inbreds using Sequence Related Amplified Polymorphism (SRAP) Markers
Himanki Dabral1,2, Ramesh Kumar Singh3, Dinesh Chandra Baskheti1, Rajeev Singh4, Arkaja Goswami5 and Anu Singh3,6*
1Department of Genetics and Plant Breeding, College of Agriculture, Govind Ballabh Pant University of Agriculture and Technology, Pantnagar, Udham Singh Nagar-263145, Uttarakhand, India
2Department of Genetics & Plant Breeding, School of Agricultural Sciences, Shri Guru Ram Rai University, Dehradun, Uttarakhand, India
3Samvetan Society for Social and Scientific Research, Dehradun-248001, Uttarakhand, India
4Materials Research Laboratory, Department of Chemistry, ARSD College, University of Delhi, New Delhi, India
5Department of Chemistry, Shyam Lal College, University of Delhi, Delhi, India
6School of Biotechnology, Jawaharlal Nehru University, New Delhi, India
- *Corresponding Author:
- Anu Singh
- School of Biotechnology, Jawaharlal Nehru University, New Delhi-110067, India
- E-mail:classicsingh@gmail.com
Received date: May 2, 2022, Manuscript No. IPJPSAR-22-13740; Editor Assigned date: May 3, 2022, PreQC No. IPJPSAR-22-13740 (PQ); Reviewed date: May 18, 2022, QC No. IPJPSAR-22-13740; Revised date: May 24, 2022, Manuscript No. IPJPSAR-22-13740 (R); Published date: May 30, 2022, DOI: 10.36648/plant-sciences.6.3.78
Citation: Dabral H, Singh RK, Baskheti DC, Singh R, Goswami A, et al. (2022) Assessment of Molecular Diversity among Maize Inbreds using Sequence Related Amplified Polymorphism (SRAP) Markers. J Plant Sci Agri Res Vol.6 No.3: 78.
Abstract
Genetic variance and association between 11 maize inbred lines were assessed using Sequence Related Amplified Polymorphism (SRAP) markers. Out of 13 SRAP marker combinations used, 11 were observed to be polymorphic obtaining a PIC value of 0.632. The assessment of Jaccard’s similarity coefficients varies between 0.69 to 0.97 among tested genotypes. Genotypes 3, and 4 (Pop 45– C8 -72-2-1-1-2 and Pob 31 23-1-1-1-1- 2-1/2# 2-2 to 6) were found to be most divergent with a similarity coefficient of 0.69. SRAP data were used to construct a dendrogram that divided the tested genotypes into two groups each having three clusters where the first subgroup (Aa) contains a single genotype G3 in Cluster II shared similarity with Cluster I with a similarity coefficient of 0.90. Between genotype G10 (Tarun 6-5-3-1-2-1-1-1) and G11 (V 116- 1) of cluster III of subgroup (Ab), the maximum similarity was found with a similarity coefficientof0.97. This work proved that the SRAP markers system is highly efficient for the identification of potential donors for the maize breeding program. Due to the use of lesser markers, a lesser number of parents with diverse origins were identified therefore more coverage of markers will be required in the future.
Keywords
Genetic variance; Maize; SRAP marker; Hybridization; Jaccard’s similarity index.
Introduction
Maize (Zea mays) belongs to the tribe, Tripsaceae (Maydeae) of the family Gramineae. It is one of the most important resourceful crops that are adapted to a wide range of agroclimatic conditions. As compared to the other crops maize is more adapted to variable environmental conditions. Being a tropical crop it requires a warm moist climate and can be grown up to an elevation more than 3000 m above sea level and require annual rainfall in the range of 250 mm to 5000 mm per year [1,2]. Due to its wider adaptability, high genetic yield potential with total world production of 1215.62 m MT among cereals, and increased industrial application demand it is known as the queen of cereals [3]. In the year 2020-2021 India produced 30 MT of maize (agricoop.nic.in). Utilization of DNA markers to identify variation or diversity in breeding lines is not only important for the improvement and speeding up the maize breeding program but also helps in the long-term conservation of this germplasm in gene banks. Several types of molecular markers have been reported to be utilized for the identification of valuable genes, and for the identification of variation at the genetic level among different genotypes of Zea mays. Sequence-Related Amplified Polymorphism (SRAP) is a new Polymerase Chain Reaction (PCR) -based molecular marker first effectively utilized in Brassica spp. by Li et al. [4]. It targets the open reading frame resulting in the formation of moderate numbers of co-dominant markers. It has a high rate of polymorphism and provides more information therefore utilized for diversity analysis both at the genetic and species level [5-8]. As compared to RAPD and AFLP markers SRAP is easily reproducible with high throughput. It has been effectively utilized in different crops like pea, lentil, and alfalfa [9,10,11,12]. In order to achieve success in the maize breeding program and for the development of heterotic maize hybrids, it is very important to explore genetic diversity and establish relationships among Z. mays germplasm. Present work proposes to find out the most potent genotypes for the development of heterotic hybrids by assessing their genetic diversity in Zea mays L., genotypes using sequence amplified polymorphic marker.
Materials and Methods
Plant materials
The genotype consisting of 11 inbred lines were collected from Vivekananda Parvatiya Krishi Anusandhan Sansthan (VPKAS), Almora, all india coordinated maize improvement programme, and some of the inbreds were taken from the university itself (Table 1). The inbred lines grew during the Kharif season at Norman E Borlaug crop research centre, at GB Pant university of agri & tech, Uttarakhand, India.
| S.No. | Genotype | Pedigree |
|---|---|---|
| 1 | G1 | TarunU 83-1-3-2-3-2-1 |
| 2 | G2 | Pob 445U -74-2-2-BBB |
| 3 | G3 | Pop 45- C8 -72-2-1-1-2U |
| 4 | G4 | Pob 31 U 23-1-1-1-1-2-1/2 # U 2-2 to6 U |
| 5 | G5 | YHP BU 45-1-2-3-1-6-2-4 U 4 |
| 6 | G6 | POB 31U 18-2-1-1-1-1-3-1 to 6 # 1-1 to 5 |
| 7 | G7 | POB 445U-58-6-3-BBB |
| 8 | G8 | POB 446U-74-2-2-BBB C8 |
| 9 | G9 | DBR N 21 |
| 10 | G10 | TarunU 6-5-3-1-2-1-1-1 |
| 11 | G11 | V 116-1 |
Table1: List of maize genotypes.
Extraction of genomic DNA
Leaves of 11 maize genotypes were used for the extraction of genomic DNA using Cetyltrimethylammonium bromide method [13]. Grounded tissue was suspended in 800 µl extraction buffer in a centrifuge tube. the centrifuge tube containing the grounded tissue was placed at 60°C in a water bath for 30 min, then, the sample was allowed to cool at room temperature, and then 600 µl of 24:1 ratio of chloroform and isoamyl alcohol was added to ensure emulsification the tubes were vortex for 10 min followed by 10 min centrifugation at 5000 rpm. Again chloroform isoamyl alcohol was added to extract the aqueous phase which was transferred to fresh tubes in which chilled isopropanol in the ratio of 3:4 by volume was added which form the precipitation of DNA CTAB complex, which was then placed at -20°C for overnight. Later, it was centrifuged at 5000 rpm at 4°C for 10 min. Finally, it was washed with chilled 70% ethanol, then again pellet was agitated and centrifuged at 5000 rpm at 4°C for 10 min and suspended in 400 µl of 1 X TE buffer after drying and stored at -20°C.
PCR amplification by SRAP marker
13 SRAP primers were used for the amplification of genomic DNA from 11 maize inbred lines using PCR thermocycler in two phases of PCR cycles. The reaction mixture to run the PCR was prepared by adding Taq polymerase enzyme as mentioned below:

SRAP primers were used in different combinations with different reaction conditions. The reaction conditions used for primers MSM1, MSM2, MSM3, and MSM4 are shown in Figure 1(a), reaction conditions used for primers MSM5, MSM6, MSM7, MSM8 and MSM9 are shown in Figure 1(b), and reaction conditions used for primers MSM10, MSM11, MSM12and MSM13 are shown in Figure 1(c). The annealing temperature in Phase 2 is different to amplify the product of interest.
Amplified DNA bands were resolved using 8% acrylamide with 42% urea as denaturing gel. The gel was supplied with 400V constant voltage for 20 min to pre-run the gel. The obtained amplified product was then added to a DNA sequencing stop solution and electrophoresis was performed for 5 h in 1X-TBE buffer at a constant voltage of 100 V and pH 8.0 after that amplified bands were visualized under UV trans-illuminator.
Primer selection
As per the primer combination suggested by Li et al., 3 forward primers were used with numbering (ME1, ME2 and ME4) and 6 reverse primers (EM1-EM6) forming 13 primer pair combination which is shown in Table 2 [3].
Analysis of molecular data
For analysis of data only polymorphic fragments that were clearly scored were used. Independent scoring of the amplified fragments was done and 0-1 matrix was formed in a spreadsheet where 1 represents fragment presence and 0 represents fragment absence. NTSYS-PC (version2.11W) was used for the analysis of binary data [14]. Jaccard’s similarity coefficients were obtained using the SIMQUAL program where genetic diversity was calculated using this formula:
Jaccard’s coefficient=NAB/(NAB+NA+NB)
Where NAB represents bands number shared by samples, NA represents sample A amplified fragments and NB denotes sample B amplified fragments. Using these indices similarity matrices were calculated. Unweighted Pair Group Method with Arithmetic Average (UPGMA) dendrogram were constructed to represent genetic diversity among the tested genotypes using similarity matrices. Using POWERMAKER software from Polymorphic Information Centre (PIC) values for each primer combination were calculated [15]. The number of the band of each SRAP locus indicates allele number. The chance that two randomly chosen alleles from a population of 11 maize inbred lines are different represents its gene Diversity (D) which was calculated using the below-given formula:
D=1-∑Pi2
Where Pi=Frequency of ith allele.
Results and Discussion
Using 3 forward and 6 reverse primers a total of 13 primer pair combinations were evaluated for their ability to prime PCR amplification of 11 inbred lines of maize. The properties of each 13 primers used to amplify 11 genotypes are presented in Table 2. A total of 99 differently sizes fragments were generated using13 primer pairs, out of which only 11 primer pairs showed polymorphism [16]. The number of SRAP fragment generated vary among different primer pair ranging from 3 in MSM10, MSM11, and MSM 13 primers to12 in MSM6 primers represented in Table 3 with an average number of 7alleles per primer. 100% polymorphic fragments were produced by MSM5, MSM7, MSM11, and MSM13 primers. Therefore, the ability of a locus to discriminate among the lines is important for the establishment of PIC values. Its value ranges from 0.0000 (primer MSM 12) to 0.8633 (primer MSM 1and primer MSM 7) (Tables 2 and 3).
| S/N | Primer | Primer code | Ends | Primer sequence 5’-3’ | Mer | % GC | °C |
|---|---|---|---|---|---|---|---|
| 1 | ME1+ EM1 | MSM 1 | Forward reverse | TGAGTCCAAACCGGATA | 17 | 53 | 52 |
| GACTGCGTACGAATTAAT | 18 | 44 | |||||
| Forward reverse | TGAGTCCAAACCGGATA | 17 | 53 | ||||
| 2 | ME1 +EM2 | MSM 2 | GACTGCGTACGAATTTGC | 18 | 56 | 52 | |
| Forward reverse | TGAGTCCAAACCGGATA | 17 | 53 | ||||
| 3 | ME1 +EM3 | MSM 3 | GACTGCGTACGAATTGAC | 18 | 47 | 52 | |
| Forward reverse | TGAGTCCAAACCGGATA | 17 | 53 | ||||
| 4 | ME1+ EM6 | MSM 4 | GACTGCGTACGAATTGCA | 18 | 47 | 52 | |
| Forward reverse | TGAGTCCAAACCGGATA | 17 | 53 | ||||
| 5 | ME1+ EM4 | MSM 5 | GACTGCGTACGAATTTGA | 18 | 47 | 50.9 | |
| Forward reverse | TGAGTCCAAACCGGATA | 17 | 53 | ||||
| 6 | ME1+ EM5 | MSM 6 | GACTGCGTACGAATTAAC | 18 | 53 | 50.9 | |
| Forward reverse | TGAGTCCAAACCGGAGC | 17 | 53 | ||||
| 7 | ME2+ EM2 | MSM 7 | GACTGCGTACGAATTTGC | 18 | 56 | 50.9 | |
| Forward reverse | TGAGTCCAAACCGGAGC | 17 | 53 | ||||
| 8 | ME2+ EM3 | MSM 8 | GACTGCGTACGAATTGAC | 18 | 47 | 50.9 | |
| Forward reverse | TGAGTCCAAACCGGAGC | 17 | 53 | ||||
| 9 | ME2+ EM6 | MSM 9 | GACTGCGTACGAATTGCA | 18 | 47 | 50.9 | |
| Forward reverse | TGAGTCCAAACCGGAGC | 17 | 53 | ||||
| 10 | ME2+ EM4 | MSM 10 | GACTGCGTACGAATTTGA | 18 | 47 | 48.8 | |
| Forward reverse | TGAGTCCAAACCGGAGC | 17 | 53 | ||||
| 11 | ME2+ EM5 | MSM 11 | GACTGCGTACGAATTAAC | 18 | 53 | 48.8 | |
| Forward reverse | TGAGTCCAAACCGGACC | 17 | 44 | ||||
| 12 | ME4+ EM4 | MSM 12 | GACTGCGTACGAATTTGA | 18 | 47 | 48.8 | |
| Forward reverse | TGAGTCCAAACCGGACC | 17 | 44 | 48.8˚ | |||
| 13 | ME4+ EM5 | MSM 13 | GACTGCGTACGAATTAAC | 18 | 53 |
Table 2: SRAP primers used for molecular diversity analysis in maize.
| Primer | No. of amplified alleles | Frequency of major alleles | Polymorphic bands | Monomorphic bands | (%) | Gene diversity | PIC |
|---|---|---|---|---|---|---|---|
| polymorphism | Value | ||||||
| MSM1 | 9 | 0.1818 | 8 | 1 | 88.9 | 0.876 | 0.8633 |
| MSM2 | 9 | 0.4545 | 7 | 2 | 77.8 | 0.7438 | 0.7213 |
| MSM3 | 7 | 0.2727 | 5 | 2 | 71.5 | 0.7934 | 0.7631 |
| MSM4 | 9 | 0.2727 | 8 | 1 | 88.9 | 0.8264 | 0.8043 |
| MSM5 | 11 | 0.2727 | 11 | 0 | 100 | 0.843 | 0.8254 |
| MSM6 | 12 | 0.3636 | 11 | 1 | 91.7 | 0.7769 | 0.7469 |
| MSM7 | 9 | 0.1818 | 9 | 0 | 100 | 0.876 | 0.8633 |
| MSM8 | 10 | 0.2727 | 9 | 1 | 90 | 0.8099 | 0.7852 |
| MSM9 | 10 | 0.3636 | 8 | 2 | 80 | 0.7769 | 0.7469 |
| MSM10 | 3 | 1 | 0 | 3 | 0 | 0 | 0 |
| MSM11 | 3 | 0.7273 | 3 | 0 | 100 | 0.4298 | 0.3855 |
| MSM12 | 4 | 1 | 0 | 3 | 0 | 0 | 0 |
| MSM13 | 3 | 0.4545 | 4 | 0 | 100 | 0.7438 | 0.7213 |
| Mean | 7.7 | 0.4476 | 6.4 | 1.3 | 76.1 | 0.6535 | 0.6328 |
Table 3: Primers used for SRAP marker and the number of polymorphic fragments revealed by each primer combination.
Therefore, based on the polymorphic nature of SRAP marker it could be utilized as an efficient tool for the identification of diverse genotypes. The lowest PIC value of (0.0000) was found to be observed for MSM 12 and MSM 1 and MSM 7 primer showed the highest PIC value of (0.8633), while SRAP amplified fragments of primers MSM7, MSM11, MSM13 were showed in Figure 2. [17].
Figure 2:PCR amplification product of 11 genotypes of maize inbred with (a) Primer MSM7, (b) MSM 11 and (c) MSM 13. The details of the genotype G1 to G11 is given in Table 1.
SRAP marker data was used to establish Jaccard’s similarity coefficients with a similarity coefficient in a range of 0.69 to 0.97 shown in (Table 4) with highest value (0.97) genetic similarity was recorded between genotype 9 (DBRN 21) and genotype 10 (Tarun 6-5-3-1-2-1-1-1), followed by genotype 1 and 2 (Tarun 83-1-3-2 and Pob 445 - 74-2-2-BBB) with a similarity coefficientof0.92. The genotypes1(Tarun 83-1-3-2), genotype 6(POB-31 18-2-1-1-1-1-3-1 to 6 # 1-1 to 5) and genotype 7 (POB 445 -58-6-3-BBB) were associated with each other with a similarity coefficient of 0.87. The similarity coefficient between pair of genotype 1,2,3,5,6 and 7 (Tarun 83- 1-3-2, Pob445-74-2-2-BBB, Pop45-C8-72-2-1-1-2 ,YHP-B 45-1-2-3-1-6-2-4 4, POB-31 18-2-1-1-1-1-3-1 to 6 # 1-1to 5 and POB 445 -58-6-3-BBBwere 0.80. The genotypes-1,3, 5(Tarun 83-1-3-2, Pop 45-C8-72-2-1-1-2 and YHP-B 45-1-2-3-1-6-2-4 4) exhibited similarity coefficient of 0.79. The lowest value for genetic similarity (0.69) was found between genotype 3and 4(Pop45-C8-72-2-1-1-2 andPob31 23-11-1-1-2-1/2 # 2-2 to 6).
| Genocy-pe | G1 | G2 | G3 | G4 | G5 | G6 | G7 | G8 | G9 | G10 | G11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | 1 | ||||||||||
| G2 | 0.92 | 1 | |||||||||
| G3 | 0.87 | 0.92 | 1 | ||||||||
| G4 | 0.80 | 0.80 | 0.85 | 1 | |||||||
| G5 | 0.79 | 0.78 | 0.79 | 0.83 | 1 | ||||||
| G6 | 0.77 | 0.77 | 0.76 | 0.74 | 0.84 | 1 | |||||
| G7 | 0.74 | 0.72 | 0.69 | 0.69 | 0.82 | 0.87 | 1 | ||||
| G8 | 0.79 | 0.76 | 0.74 | 0.73 | 0.80 | 0.88 | 0.87 | 1 | |||
| G9 | 0.82 | 0.81 | 0.80 | 0.78 | 0.79 | 0.80 | 0.82 | 0.84 | 1 | ||
| G11 | 0.83 | 0.83 | 0.80 | 0.76 | 0.78 | 0.82 | 0.80 | 0.85 | 0.97 | 1 | |
| G12 | 0.81 | 0.84 | 0.80 | 0.75 | 0.79 | 0.81 | 0.82 | 0.83 | 0.94 | 0.97 | 1 |
Table 4: The Jaccard’s similarity coefficients were estimated between pairs of inbred lines on the basis of SRAP markers.
Genetic diversity among inbred lines
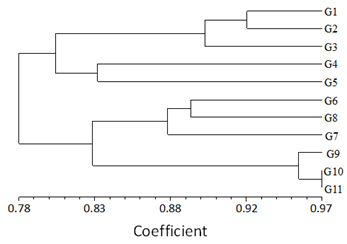
The objective of using SRAP markers was to estimate the level of genetic diversity among different genotypes. Jaccard’s similarity coefficients based on the generated data by SRAP marker were used to develop UPGMA mean dendrogram (Figure 3). The UPGMA ordered the population of 11 inbred lines into one main group (Group A) which is divided into 2 subgroups (sub Aa) and a subgroup (sub-Ab). Subgroup Aa is further subdivided into 3 clusters (I, II, and III). cluster I comprised 2 genotypes G1 and G2 which have a similarity coefficient value of 0.92. A similarity coefficient of 0.90 was shared between cluster I and cluster II consisting of genotype G3. Similarly, cluster III is comprised of 2 genotypes G4 and G5, and the similarity coefficient between them is 0.83. Further subgroup (sub-Ab) is divided into 3 clusters (I, I, and III). Cluster I is comprised of 2 genotypes G6 and G8 with a similarity coefficient value of 0.89 and cluster II comprises of single genotype G7 which is sharing a similarity coefficient value of 0.88 with cluster I, and lastly, cluster III comprises of 3 genotypes G9, G10 and G11. Genotypes G10 and G11 have a similarity coefficient value of 0.97. Clusters II and III show a similarity coefficient of 0.83.
Conclusion
The presence of high genetic variability with a broad genetic base was found to be observed with 1a 00% polymorphic rate in our collection as shown by (MSM5, MSM7, MSM11, and MSM13) primers. This marker was found to be the most effective and efficient tool for testing the genetic polymorphism and relationship among the tested maize genotypes with the production of a large number of polymorphic bands with an average of 7.62 alleles per primer detected. This information will be useful in the future for designing breeding strategies by making use of such large allelic variations that can be used to develop desirable gene combinations in the maize improvement program.
Acknowledgements
HD is thankful to all the technical support provided by the department. AS acknowledges the funding from UGC (F.15-1/2016-17/PDFWM-2015-17-UTT-33566(SA-II)).
Additional Information
The second author has contributed equally to the research findings.
References
- Corn and corn improvement (1988)Climatic requirement. In: Sprague GF,Dubey JW (eds.) American Society of Agronomy, Madison, USA, pp: 609-616.
- Dowswel CR, Cantrell RP (1996) Maize in the third world. West view press USA, pp. 43-55.
[Crossref], [Google Scholar]
- USDA 2022. Data and statistics.
- Li GY, Quiros CF (2001) Sequence-Related Amplified Polymorphism (SRAP), a new marker system based on a simple PCR reaction, its application to mapping and gene tagging in Brassica. Theor Appl Genet 103: 455-461.
[Crossref], [Google Scholar]
- Wang X, Liu G, Chang R, Han J, Guo E (2009) Optimization of annealing temperature of SRAP-PCR in 5 temperate fruits. Genom Applied Biol 28: 525-528. [Crossref],
- Riaz A, Quresh Li, Swati MS, Quiros CF (2001) Genetic diversity of oilseed Brassica napusinbred lines based on sequenceâ€Ârelated amplified polymorphism and its relation to hybrid performance. Plant Breeding 120: 411-415.
[Crossref], [Google Scholar], [Indexed]
- Budak H, Shearman RC, Parmaksiz I, Dweikat I (2004) Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theor Appl Genet 109: 280-288.
[Crossref], [Google Scholar], [Indexed]
- Lin ZX, Zhang XL, Nie YC (2004) Evaluation of application of a new molecular marker SRAP on analysisof F2 segregation population and genetic diversity in cotton. Acta Genetica Sinica 3: 1622-626.
[Crossref], [Google Scholar], [Indexed]
- Vandemark GJ, Ariss JJ, Bauchan GA, Larsen RC, Hughes TJ (2006) Estimating genetic relationships among historical sources of alfalfa germplasm and selected cultivars with sequence related amplified polymorphisms. Euphytica 152: 9-16. [Crossref],
[Google Scholar], [Indexed]
- Ariss JJ, Vandemark GJ (2007) Assessment of genetic diversity among non dormant and semidormant alfalfa populations using sequence-related amplified polymorphisms. Crop Sci 47: 2274-2284.
[Crossref], [Google Scholar], [Indexed]
- Castonguay Y, Cloutier J, Bertrand A, Michaud R, Laberge S (2010) SRAP polymorphisms associated with superior freezing tolerance in alfalfa (Medicagosativaspp. sativa). Theor Appl Genet 120: 1611-1619.
[Crossref], [Google Scholar], [Indexed]
- Esposito MA, Martin EA, Craverom VP, Cointry E (2007) Characterization of pea accessions by SRAP’s markers. Sci Hortic 113: 329-335.
[Crossref], [Google Scholar],
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13-15.
- Rohlf FG (1993) NTSYS-pc. Numerical taxonomy and multivariate analysis system.2, Applied Biostatistics, New York.
- Liu K, Muse S (2005) Power marker: An integrative analysis environment for genetic marker analysis. Bioinformatics 21: 2128-2129.
[Crossref], [Google Scholar], [Indexed]
- Bhat J, Kumar S, Patel S, Solanki R (2017) Sequence-related amplified polymorphism (SRAP) markers based genetic diversity analysis of cumin genotypes. Ann Agrar Sci 15: 434-438.
[Crossref], [Google Scholar], [Indexed]
- Abdelkhalik SM, Salem AKM, Abdelaziz AR, Ammar MH (2016) Morphological and sequence-related amplified polymorphism-based molecular diversity of local and exotic wheat genotypes. Genet Mol Res 15: 1-9.
[Crossref], [Google Scholar], [Indexed]
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences