Almespar: An Open Reading Frames Detection Tool Using Python Programing Language
Osamah S Alrouwab1,2*, Buthaynah H Ramadhan2, Kareemah A Abdullah2, Omiema M Aznad2, Saifedden Ayad3 and Mahmoud Gargotti4
1Department of Biochemistry, Alzintan University, Gharbi, Libya
2Department of Biotechnology, Aljafra University, Alsahla, Libya
3Department of Preventive Medicine, Al-Zaytoonah University, Tarhuna, Libya
4Department of Microbiology, University of Zawia, Zawia, Libya
- *Corresponding Author:
- Osamah S Alrouwab
Department of Biochemistry,
Alzintan University,
Gharbi,
Libya;
E-mail: usamaerawab@gmail.com
Received date: December 14, 2022, Manuscript No. IPMGM-22-15373; Editor assigned date: December 19, 2022, PreQC No. IPMGM-22-15373 (PQ); Reviewed date: January 03, 2023, QC No. IPMGM-22-15373; Revised date: March 20, 2023, Manuscript No. IPMGM-22-15373 (R); Published date: March 28, 2023, DOI: 10.36648/ IPMGM.7.1.001
Citation: Alrouwab OS, Ramadhan BH, A bdullah KA, A znad OM, Ayad S , et a l. ( 2023) A lmespar: A n O pen Reading Frames Detection Tool Using Python Programing Language. J Mol Genet Med Vol:7 No:1
Abstract
Open Reading Frames (ORFs) are sections of a reading frame that do not include any stop codons. A reading frame is a sequence of nucleotide triplets read as codons indicating amino acids; a single strand of DNA has three potential reading frames. Long ORFs in a DNA sequence may represent possible protein coding areas. In addition to extended ORFs, which assist in gene locus prediction, there is yet another type of ORFS known as small Open Reading Frames (smORFs), which have 100 codons or fewer. The fundamental purpose of this project is to develop an offline, cross-platform, and dependable detection tool for regular ORFs and smORFs prevalent in biomedical studies. In this work, the most ORFs were found in the Bos taurus (cattle) insulin gene, which had 17 consecutive ORFs, while the fewest ORFs were reported in the Cani's lupus (dog) insulin gene, which had only 4 ORFs. In general, the software meets the expected demarcation restrictions. We strongly advise more research into the detection of nested ORFs.
Keywords
Open Reading Frames (ORFs); small Open Reading Frames (smORFs); Nested ORFs; Codons; Insulin gene
Introduction
Molecular biology introduces the Open Reading Frames (ORFs) as stretches of DNA sequence between the start and stop codons [1]. Rapid and accurate identification of all conceivable ORFs from DNA Sequence with known genetic coding appears to be an unpretentious procedure which may be accomplished online at, in practice, this process is hampered by DNA sequencing mistakes, which may result in the omission or incorrect assignment of start/stop codons, resulting in longer or truncated ORFs. When confronted with a list of all potential ORFs in a genome, determining which ones comprise genes can be challenging. To begin, substantially or completely overlapping ORFs frequently coexist on the same DNA strand. Second, conflicting ORFs are frequently found on distinct DNA strands. Finally, even if no conflicts exist, there is no guarantee that an ORF, specifically a short one, genuinely translates for a protein. Numerous genes have been identified that express transcripts with mRNA like properties, such as capping and polyadenylation, although they do not appear to be translated into proteins; such transcripts are referred as long non-coding RNAs (lncRNAs). Those genes and their byproducts have fundamentally altered our knowledge of transcriptional regulation. Additional form of gene elements complicates our understanding of the genome's coding potential: small ORFs (smORFs; occasionally known as sORFs) with 10 to 100 codons that are thought to be functional. There are huge numbers of smORF sequences in nucleotide sequences, and several of them may be linked to transcripts, and in many cases, to presumed lncRNAs. As a result, useable smORFs are frequently not annotated because they have still not been empirically verified, and they have not been confirmed because they are not annotated, a challenge that is seldom (and only by chance) surmounted. The issue with algorithmic annotation is that, like canonical protein-coding ORFs, it is based completely on sequence similarities, which disclose the conservation of the presumed coding sequence, denoting a selective result and thus function; and resemblance to proteins and protein domains with an observationally substantiated function, indicating a comparable performance for the smORF [2]. Genes are commonly detected on the basis of statistically considerable resemblance between translated ORFs and recognized gene products. Gene identification approaches based on coding potential evaluation and detection of regulatory DNA elements must be used in the absence of authentic datasets. Identifying ORFs is critical in biochemical and molecular practice. Despite of the abundance of ORF detection tools available, most of them were web based and demanded an internet connection. As a result, the primary goal of this study is to provide an offline, cross-platform, and reliable ORFs detection tool for regular implementation in biological research.
Materials and Methods
Data mining
The datasets used to assess the application's efficiency as well as the formulation of probabilistic scenarios, were collected from online, publicly available databases, namely the GenBank databases from The National Center for Biotechnology Information, on January 18, 2021 [3]. Five distinct whole genome shotgun sequences of the INS (Insulin) gene were utilized to characterize various Mammalian species:
• Bos taurus; cattle (accession ID: NC_037356.1),
• Canis lupus; dog (accession ID: NC_051822.1),
• Felis catus; domestic cat (accession ID: NC_058377.1),
• Homo sapiens; human (accession ID: NC_000011.10),
• Sus scrofa; pig (accession ID: NC_010444.4).
The genetic codes matrix
The codons database was obtained from NCBI taxonomy database, on March 23, 2021 to build the search patterns matrix. NCBI takes considerable effort to guarantee that every Coding Sequence (CDS) in GenBank data has the correct translation [4]. The meticulous validation of each record's taxonomy and assignment of the right genetic code for each organism and record is key to this endeavor (Table 1).
| Genetic code in taxonomy tree | Initiation codons | Stop codons |
|---|---|---|
| The standard code | TTG, CTG, ATG | TAA, TAG, TGA |
| The vertebrate mitochondrial code | ATT, ATC, ATA, ATG, GTG | TAA, TAG, AGA, AGG |
| The yeast mitochondrial code | ATA, ATG | TAA, TAG |
| The mold, protozoan and coelenterate mitochondrial code | TTA, TTG, CTG, ATT, ATC, ATA, ATG, GTG | TAA, TAG |
| The invertebrate mitochondrial code | TTG, ATT, ATC, ATA, ATG, GTG | TAA, TAG |
| The ciliate, dasycladacean and hexamita nuclear code | ATG | TGA |
| The echinoderm and flatworm mitochondrial code | ATG, GTG | TAA, TAG |
| The euplotid nuclear code | ATG | TAA, TAG |
| The bacterial, archaeal and plant plastid code | TTG, CTG, ATT, ATC, ATA, ATG, GTG | TAA, TAG, TGA |
| The alternative yeast nuclear code | CTG, ATG | TAA, TAG, TGA |
| The ascidian mitochondrial code | TTG, ATA, ATG, GTG | TAA, TAA, TAG |
| The alternative flatworm mitochondrial code | ATG | TAG |
| Chlorophycean mitochondrial code | ATG | TAA, TAG |
| Trematode mitochondrial code | ATG, GTG | TAA, TAG |
| Scenedesmus obliquus mitochondrial code | ATG | TCA, TAA, TGA |
| Thraustochytrium mitochondrial code | ATT, ATG, GTG | TTA, TAA, TAG, TGA |
| Rhabdopleuridae mitochondrial code | TTG, CTG, ATG, GTG | TAA, TAG |
| Candidate division SR1 and gracilibacteria code | TTG, ATG, GTG | TAA, TAG |
| Pachysolen tannophilus nuclear code | CTG, ATG | TAA, TAG, TGA |
| Karyorelict nuclear code | ATG | TGA |
| Condylostoma nuclear code | ATG | TAA, TAG, TGA |
| Mesodinium nuclear code | ATG | TGA |
| Peritrich nuclear code | ATG | TGA |
| Blastocrithidia nuclear code | ATG | TAA, TAG, TGA |
| Cephalodiscidae mitochondrial UAA-Tyr code | ATG | TAA, TAG, TGA |
Table 1: Genetic code in taxonomy tree.
Implementation
The benchmarks were all completed using an Intel (R) Core (TM) i5-3470 CPU running at 3.20 GHz and 16 GB of DDR3 RAM. Ubuntu Linux Desktop 20.04 LTS/ 64-bit was utilized as the operating system for the benchmarks [5]. Python programming language version 3.9.5 was used to develop the application.
Design
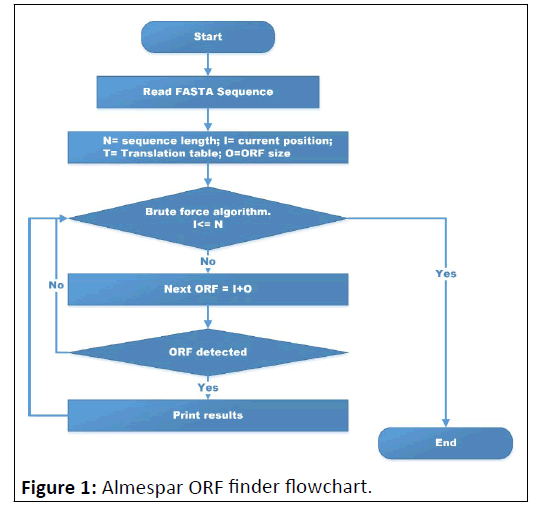
To determine the initialization and stop codons, a modified form of a brute-force algorithm for exact string matching was recruited, the new search begins from the last successful stop codon, so that the ORFs overlapping in this approach cannot be detected, which considered as one of the methodology's shortcomings (Figure 1).
Results and Discussion
This project aims to build Almespar, cross-platform offline software for locating Open Reading Frames (ORFs) over different species. The Insulin (INS) gene was utilized as a target in this study to assess the program's reliability in identifying open reading frames in five mammal species [6]. The rediscovery of insulin signifies a genuine milestone, highlighted by contrasts, arguments, and disagreements among experts, and perhaps even significant frustrations, setbacks, and occasionally optimism. The advent of insulin was a watershed moment in diabetes diagnosis and treatment, radically revolutionizing both therapy and prognosis. Diabetes is one of the most researched disorders in medical history, with the oldest mentions dating back to a collection of Egyptian medical scripts written near 1552 BC, known as the Ebers Papyrus.
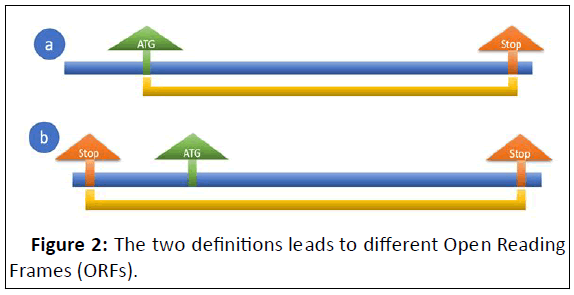
Insulin is a crucial hormonal modulator of development and metabolism in mammals and may have a comparable role in many other eukaryotes, while clear structural evidence on insulin like molecules found outside of vertebrates is still absent [7]. In the lack of insulin, many cells in the body fail to use glucose and amino acids correctly, resulting in severe metabolic derangements [8-10]. In man, inability to metabolize glucose results in diabetes mellitus, which is characterized by glucosuria, ketonuria, growth arrest, and negative nitrogen balance, eventually leading to death from either acute metabolic acidosis caused by unrestrained fatty acid oxidation or, in the absence of sufficient lipid stores to generate ketone bodies, from inanition hence the classic description of the body "melting down into urine" in diabetes [11]. The terminology Open Reading Frame (ORF) is fundamental in gene discovery. Interestingly, two concepts are being used. An ORF is described in all definitions as a span of nucleotide sequence that is not disrupted by stop codons in a specific reading frame, although they diverge in the follows:
• An ORF is a sequence with a distance that is divided by three letter which starts with a translation start codon (ATG) and terminates with a stop codon as illustrated in Figure 2a,
• An ORF is a sequence with a length that is divisible by three letter and is delimited by stop codons as shown in Figure 2b [12,13].
ORFs in Bos taurus (cattle) INS gene
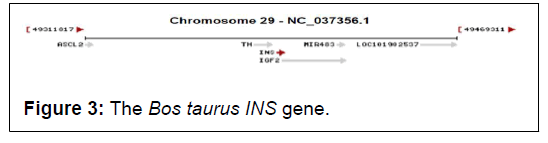
The Bos taurus INS gene (Gene ID: 280829) spans 1620 pb on chromosome 29 (Figure 3).
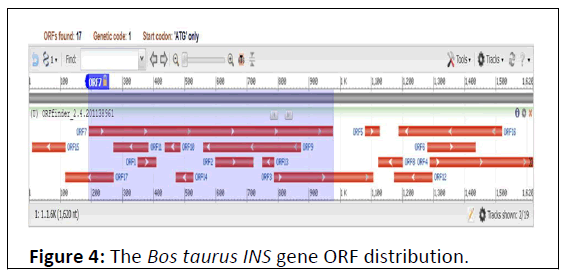
The number of ORF’s mined were 17 spreads across the gene (Figure 4). The total number of forward ORFs observed was 7, ranged from 60 pb to 786 bp in length. While the lengths of the ORFs detected on the reverse strand were 39 bp to 333 bp (Table 2).
| Label | Strand | Frame | Start | Stop | Length (bp) |
|---|---|---|---|---|---|
| ORF1 | + | 1 | 349 | 488 | 60 |
| ORF2 | + | 1 | 598 | 720 | 123 |
| ORF3 | + | 1 | 787 | 1107 | 321 |
| ORF4 | + | 1 | 1288 | 1620 | 333 |
| ORF5 | + | 2 | 1079 | 1126 | 48 |
| ORF6 | + | 2 | 1280 | 1435 | 156 |
| ORF7 | + | 3 | 192 | 977 | 786 |
| ORF8 | - | 1 | 1200 | 1123 | 78 |
| ORF9 | - | 1 | 873 | 559 | 315 |
| ORF10 | - | 1 | 486 | 436 | 51 |
| ORF11 | - | 1 | 384 | 271 | 114 |
| ORF12 | - | 2 | 1295 | 1173 | 123 |
| ORF13 | - | 2 | 788 | 750 | 39 |
| ORF14 | - | 2 | 527 | 471 | 57 |
| ORF15 | - | 2 | 116 | 9 | 108 |
| ORF16 | - | 3 | 1519 | 1187 | 333 |
| ORF17 | - | 3 | 271 | 116 | 156 |
Table 2: Bos taurus (cattle) Insulin gene ORF’s.
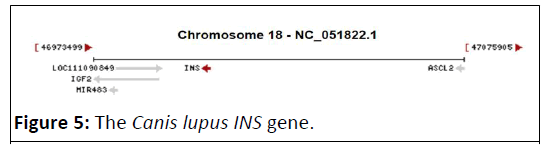
ORFs in Canis lupus (dog) INS gene
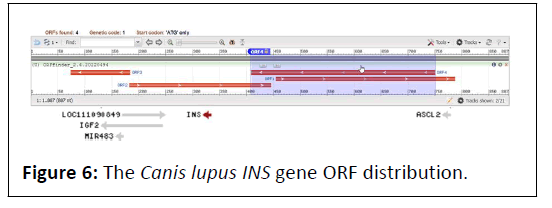
The dog, Canis lupus INS gene (Figure 5), revels a limited handful of ORFs in both strands (Figure 6) [14]. Only two ORFs detected on forward strand ranging in length from 264 bp to 333 bp, and two ORFs for the reverse strand were 111 and 342 bp long, respectively (Table 3) [15,16].
| Label | Strand | Frame | Start | Stop | Length (bp) |
|---|---|---|---|---|---|
| ORF1 | + | 2 | 455 | 787 | 333 |
| ORF2 | + | 3 | 183 | 446 | 264 |
| ORF3 | - | 2 | 184 | 74 | 111 |
| ORF4 | - | 3 | 750 | 489 | 342 |
Table 3: Canis lupus (dog) Insulin gene ORF’s.
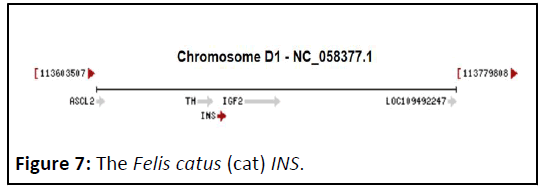
ORFs in Felis catus (cat) INS gene
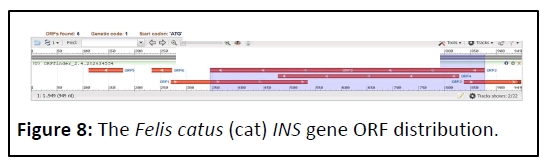
The Felis catus (cat) INS gene (Gene ID: 493804) contains only 6 ORFs (Figure 7) dispersed on both strands (Figure 8).
The total number of forward ORFs found was two, with lengths ranging from 111 pb to 264 bp [17,18]. The lengths of the ORFs discovered on the reverse strand were 4 ranged from 39 to 531 bp (Table 4).
| Label | Strand | Frame | Start | Stop | Length (bp) |
|---|---|---|---|---|---|
| ORF1 | + | 1 | 271 | 534 | 264 |
| ORF2 | + | 1 | 838 | 948 | 111 |
| ORF3 | - | 1 | 877 | 347 | 531 |
| ORF4 | - | 2 | 828 | 478 | 351 |
| ORF5 | - | 2 | 177 | 112 | 66 |
| ORF6 | - | 3 | 272 | 234 | 39 |
Table 4: Felis catus (cat) Insulin gene ORF’s.
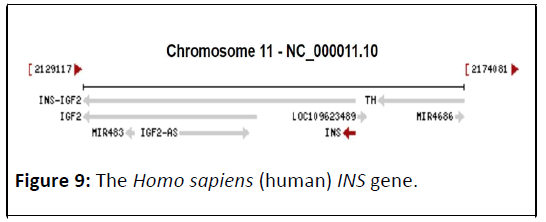
The Homo sapiens (human) INS gene
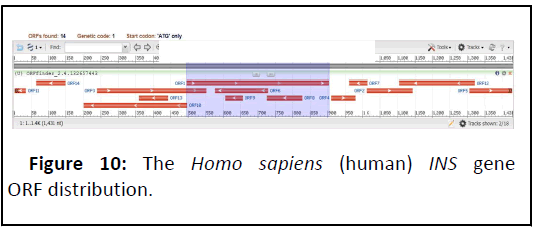
The Homo sapiens (human) INS gene (Gene ID: 3630), Its cytogenetic position Ch38.p13, and composed of 1431 DNA bp (Figure 9) [19]. It’s comprised of 14 ORFs, disseminate on both strands (Figure 10). The total ORFs number on forward strand were 5, ranging from 69 bp to 408 bp, while the reverse strand shows 9 ORFs ranging from 33 bp to 297 bp (Table 5).
| Label | Strand | Frame | Start | Stop | Length (bp) |
|---|---|---|---|---|---|
| ORF1 | + | 1 | 496 | 903 | 408 |
| ORF2 | + | 1 | 1812 | 1143 | 132 |
| ORF3 | + | 2 | 239 | 553 | 315 |
| ORF4 | + | 2 | 911 | 979 | 69 |
| ORF5 | + | 2 | 1387 | 1429 | 123 |
| ORF6 | - | 1 | 729 | 577 | 153 |
| ORF7 | - | 2 | 1013 | 963 | 51 |
| ORF8 | - | 2 | 827 | 726 | 102 |
| ORF9 | - | 2 | 656 | 606 | 51 |
| ORF10 | - | 2 | 497 | 201 | 297 |
| ORF11 | - | 2 | 35 | 3 | 33 |
| ORF12 | - | 3 | 1321 | 1106 | 216 |
| ORF13 | - | 3 | 442 | 359 | 84 |
| ORF14 | - | 3 | 148 | 65 | 84 |
Table 5: Homo sapiens (human) Insulin gene ORF’s.
ORFs in Sus scrofa (pig) INS gene
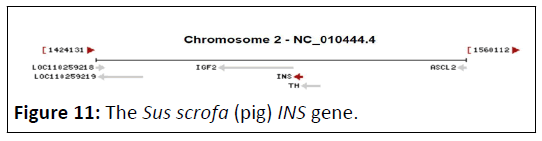
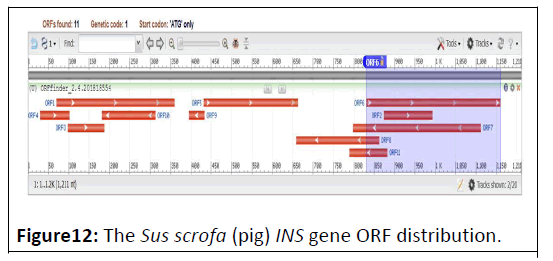
The Sus scrofa (pig) INS gene (GeneID:397415), located on chromosome 2 and composed of 1211 DNA bp (Figure 11). It’s comprised of 11 ORFs, distributed on both strands (Figure 12) [20]. The total ORFs number on forward strand were 6, ranging from 72 bp to 330 bp, while the reverse strand shows 5 ORFs ranging from 39 bp to 315 bp (Table 6).
| Label | Strand | Frame | Start | Stop | Length (bp) |
|---|---|---|---|---|---|
| ORF1 | + | 1 | 70 | 360 | 291 |
| ORF2 | + | 1 | 874 | 993 | 120 |
| ORF3 | + | 2 | 98 | 187 | 90 |
| ORF4 | + | 3 | 30 | 101 | 72 |
| ORF5 | + | 3 | 432 | 662 | 231 |
| ORF6 | + | 3 | 831 | 1160 | 330 |
| ORF7 | - | 1 | 1112 | 798 | 315 |
| ORF8 | - | 2 | 862 | 659 | 204 |
| ORF9 | - | 2 | 433 | 395 | 39 |
| ORF10 | - | 2 | 313 | 182 | 132 |
| ORF11 | - | 3 | 882 | 790 | 93 |
Table 6: The Sus scrofa (pig) Insulin gene ORF’s.
Conclusion
In this study, the highest ORFs were reported in Bos taurus (cattle) Insulin gene which scored 17 successive ORFs whereas the lowest score was reported in Cani’s lupus (dog) insulin gene which shows only 4 ORFs. Generally, the program fulfills the boundary limits as expected. We strongly recommend further work, consider detection of nested ORFs.
References
- Sieber P, Platzer M, Schuster S (2018) The definition of open reading frame revisited. Trends Genet 34:167–170
[Crossref] [Google Scholar] [PubMed]
- Ramos-Gonzalez PL, Santos GF dos, Chabi-Jesus C, Harakava R, Kitajima EW, et al. (2020) Passion fruit green spot virus genome harbors a new orphan ORF and highlights the flexibility of the 5′-end of the RNA2 segment across cileviruses. Front Microbiol 11:206
- Patraquim P, Mumtaz MAS, Pueyo JI, Aspden JL, Couso JP (2020) Developmental regulation of canonical and small ORF translation from mRNAs. Genome Biol 21:1–26
[Crossref] [Google Scholar] [PubMed]
- Stringer A, Smith C, Mangano K, Wade JT (2021) Identification of novel translated small ORFs in Escherichia coli using complementary ribosome profiling approaches. J Bacteriol 204:JB0035221
[Crossref] [Google Scholar] [PubMed]
- Dvorkina T, Bankevich A, Sorokin A, Yang F, Adu-Oppong B, et al. (2021) ORFograph: Search for novel insecticidal protein genes in genomic and metagenomic assembly graphs. Microbiome 9:1–14
[Crossref] [Google Scholar] [PubMed]
- Bartholomaus A, Kolte B, Mustafayeva A, Goebel I, Fuchs S, et al. (2021) smORFer: A modular algorithm to detect small ORFs in prokaryotes. Nucleic Acids Res 49:e89–e89
- Brunet MA, Levesque SA, Hunting DJ, Cohen AA, Roucou X (2018) Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Res 28:609–624
[Crossref] [Google Scholar] [PubMed]
- Wright BW, Molloy MP, Jaschke PR (2021) Overlapping genes in natural and engineered genomes. Nat Rev Genet 23:154–168
[Crossref] [Google Scholar] [PubMed]
- Todd RT, Wikoff TD, Forche A, Selmecki A (2019) Genome plasticity in Candida albicans is driven by long repeat sequences. Elife 8:e45954
[Crossref] [Google Scholar] [PubMed]
- Goustin AS, Thepsuwan P, Kosir MA, Lipovich L (2019) The Growth Arrest Specific (GAS)-5 long non-coding RNA: A fascinating lncRNA widely expressed in cancers. Non Coding RNA 5:46
[Crossref] [Google Scholar] [PubMed]
- Choudhary S, Li W, Smith A (2020) Accurate detection of short and long active ORFs using Ribo-seq data. Bioinformatics 36:2053–2059
[Crossref] [Google Scholar] [PubMed]
- Ransohoff JD, Wei Y, Khavari PA (2018) The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19:143–157
[Crossref] [Google Scholar] [PubMed]
- Guo CJ, Xu G, Chen LL (2020) Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci 45:947–960
[Crossref] [Google Scholar] [PubMed]
- Zimmer-Bensch G (2019) Emerging roles of long non-coding RNAs as drivers of brain evolution. Cells 8:1399
[Crossref] [Google Scholar] [PubMed]
- Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22:96–118
[Crossref] [Google Scholar] [PubMed]
- Claridge B, Kastaniegaard K, Stensballe A, Greening DW (2019) Post-translational and transcriptional dynamics regulating extracellular vesicle biology. Expert Rev Proteomics 16:17–31
[Crossref] [Google Scholar] [PubMed]
- Basrai MA, Hieter P, Boeke JD (1997) Small open reading frames: Beautiful needles in the haystack. Genome Res 7:768–771
[Crossref] [Google Scholar] [PubMed]
- Yazhini A (2018) Small open reading frames. Resonance 23:57–67
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, et al. (2006) Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38:149–153
[Crossref] [Google Scholar] [PubMed]
- Chekulaeva M, Rajewsky N (2019) Roles of long noncoding RNAs and circular RNAs in translation. Cold Spring Harb Perspect Biol 11:a032680
[Crossref] [Google Scholar] [PubMed]
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences