ISSN : 2349-3917
American Journal of Computer Science and Information Technology
A Proposal for a New Biological Supercomputer based Exclusively on RNA Molecules, Quantum Mechanics and Nanoptics
Karim Massoud* Mary Popova, John F Williams and Patricia Hernandez
Department of Software Engineering, Harper Adams University, Edgmond, United Kingdom
- *Corresponding Author:
- Karim Massoud
Department of Engineering,
Harper Adams University, Edgmond,
United Kingdom
E-mail: karimmassoud54@gmail.com
Received Date: October 12, 2020; Accepted Date: October 26, 2020; Published Date: November 02, 2020
Citation: Massoud K, Popova M, Williams JF, Hernandez P (2020) A Proposal for a New Biological Supercomputer based Exclusively on RNA Molecules, Quantum Mechanics and Nanoptics Vol.8 No.4: 63.
Abstract
In this paper, we propose a biological supercomputer which is based in RNA molecules. Biological computers are made from living cells. Instead of electrical wiring and signaling, biological computers use chemical inputs and other biologically derived molecules such as proteins and DNA. In this paper we construct a biological supercomputer which is based in RNA molecules using Quantum Mechanics and Nanoptics. We use simulation with Neural Networks and Fuzzy Logic. The theoretical results are supported by numerical analysis and two experiments.
Keywords
Super computing; Biological supercomputers; RNA; DNA; Quantum mechanics; Nanoptics
Introduction
Just like a desktop computer, these organic computers can respond to data and process it, albeit in a rudimentary manner similar to the capabilities of computers circa 1920 [1]. While biological computers have a long way to go before they are as sophisticated as today's personal computers, the fact that researchers have been able to get biological computers to complete a logic gate is a notable achievement. Once you’ve programmed a single biological cell, it’s extremely cost-effective to grow billions more with only the cost of the nutrient solutions and a lab tech’s time. It’s also anticipated that bio computers might actually be more reliable than their electronic counterparts. To illustrate, think about how our bodies still survive even though millions of our cells die off, but a computer built from wires can stop functioning if one wire is severed [2]. In addition, every cell has a mini-factory at its disposal, so once it's been programmed; it can synthesize any biological chemical. Instead of what's done today when bioengineers map genes and try to uncover their secrets, they can just program cells to do the job they need them to do for example, program cells to fight cancer or deliver insulin to a diabetic's bloodstream. Although biocomputing has similarities with biology and computer science, it doesn’t fit seamlessly with either one. In biology, the goal is to reverse engineer things that have already been built. Biocomputing aims to forward engineer biology. Experts in computer science are accustomed to machines executing programmed commands; when dealing with biological environments in what is known as “wet lab” organisms might react unpredictably. The culprit could be the cell's programming, or it could easily be something external such as the environmental conditions, nutrition, or timing. While biological computers aren’t as prolific as personal computers, there are several companies working to advance this very young field [3]. The founders of Synthego, a Silicon Valley startup, aren’t biologists. They are brothers and software engineers who used to work for Space X building rockets but thought there was potential in taking what they knew about agile design to gene-editing tools. The company creates customized CRISPR kits for scientists from a selection of approximately 5,000 organisms available in Synthego’s genome library. Ultimately, this can cut down the time it takes for scientists to do gene edits [4].
Materials and Methods
Microsoft’s foray into biological computing is called Station B. The company partnered with Princeton University and two UK companies, Oxford Bio Medica and Synthace, on the new research system that can analyze volumes of biomedical data with a set of integrated computer programs. This analysis is then used to guide scientists on the best way to proceed with research, such as editing DNA in a certain way. The hope is that this system will ultimately lower the cost of gene-therapy products to bring them too many more patients. Using CRISPR (DNA sequences found within e.g. bacteria), scientists were able to turn a cell into a biological computer [5]. It was programmed to take in specific genetic codes and perform computations that would produce a particular protein. This milestone could eventually lead to having powerful computers in cells that could eventually detect and treat diseases. Imagine in the future that these cells could be programmed to scan for biomarkers that indicate the presence of disease [6]. If all criteria are met, these same cells could massproduce proteins that could help treat the disease. A micro tissue might have billions of cells, all with their own "dual-core processor." The computing power this would allow is on par with today's digital supercomputer. The work in biocomputing thus far has focused on DNA-based systems because, at this point, genetic engineering is understood enough (even if all of its secrets aren't known) to make progress possible. There are many more biological systems to tackle, such as those based on nerve cells. The future is expected to include using the knowledge gleaned from developing biocomputers for DNA-based systems and apply it to neurochemistry. Thank you for reading my post [7]. Here at LinkedIn and at Forbes I regularly write about management and technology trends. I have also written a new book about AI, click here for more information. To read my future posts simply join my network here or click 'Follow'. Also feel free to connect with me via Twitter, Facebook, Instagram, Slideshare or YouTube. Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and DNA are nucleic acids [8]. Along with lipids, proteins, and carbohydrates, nucleic acids constitute one of the four major macromolecules essential for all known forms of life. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA, RNA is found in nature as a single strand folded onto itself, rather than a paired double strand. Cellular organisms use messenger RNA (mRNA) to convey genetic information (using the nitrogenous bases of guanine, uracil, adenine, and cytosine, denoted by the letters G, U, A, and C) that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome [9]. Some RNA molecules play an active role within cells by catalyzing biological reactions, controlling gene expression, or sensing and communicating responses to cellular signals. One of these active processes is protein synthesis, a universal function in which RNA molecules direct the synthesis of proteins on ribosomes. This process uses transfer RNA (tRNA) molecules to deliver amino acids to the ribosome, where ribosomal RNA (rRNA) then links amino acids together to form coded proteins [10].
RNA computing
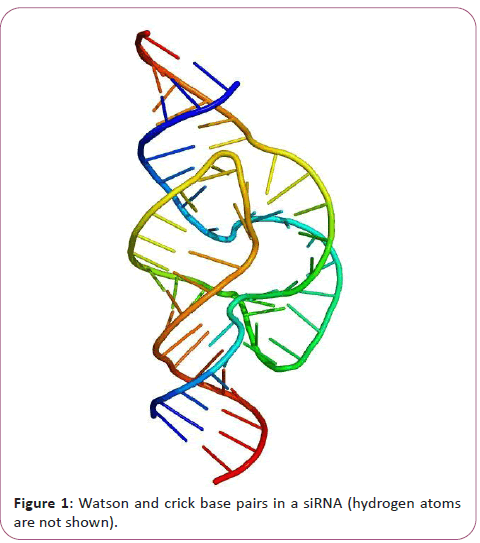
The chemical structure of RNA is very similar to that of DNA, but differs in three primary ways. Unlike double-stranded DNA, RNA is a single-stranded molecule in many of its biological roles and consists of much shorter chains of nucleotides. However, a single RNA molecule can, by complementary base pairing, form intrastrand double helixes, as in tRNA. While the sugar-phosphate "backbone" of DNA contains deoxyribose, RNA contains ribose instead. Ribose has a hydroxyl group attached to the pentose ring in the 2' position, whereas deoxyribose does not. The hydroxyl groups in the ribose backbone make RNA more chemically labile than DNA by lowering the activation energy of hydrolysis. The complementary base to adenine in DNA is thymine, whereas in RNA, it is uracil, which is an unmethylated form of thymine. Like DNA, most biologically active RNAs, including mRNA, tRNA, rRNA, snRNAs, and other non-coding RNAs, contain self-complementary sequences that allow parts of the RNA to fold and pair with itself to form double helices [11]. Analysis of these RNAs has revealed that they are highly structured. Unlike DNA, their structures do not consist of long double helices, but rather collections of short helices packed together into structures akin to proteins [12]. In this fashion, RNAs can achieve chemical catalysis (like enzymes). For instance, determination of the structure of the ribosome an RNA-protein complex that catalyzes peptide bond formation revealed that its active site is composed entirely of RNA. The chemical structure of RNA is very similar to that of DNA, but differs in three primary ways: Unlike double-stranded DNA, RNA is a single-stranded molecule in many of its biological roles and consists of much shorter chains of nucleotides [13]. However, a single RNA molecule can, by complementary base pairing, form intrastrand double helixes, as in tRNA (Figure 1).
While the sugar-phosphate "backbone" of DNA contains deoxyribose, RNA contains ribose instead. Ribose has a hydroxyl group attached to the pentose ring in the 2' position, whereas deoxyribose does not. The hydroxyl groups in the ribose backbone make RNA more chemically labile than DNA by lowering the activation energy of hydrolysis. The complementary base to adenine in DNA is thymine, whereas in RNA, it is uracil, which is an unmethylated form of thymine [14]. Like DNA, most biologically active RNAs, including mRNA, tRNA, rRNA, snRNAs, and other non-coding RNAs, contain self-complementary sequences that allow parts of the RNA to fold and pair with itself to form double helices. Analysis of these RNAs has revealed that they are highly structured. Unlike DNA, their structures do not consist of long double helices, but rather collections of short helices packed together into structures akin to proteins. In this fashion, RNAs can achieve chemical catalysis (like enzymes) [15]. For instance, determination of the structure of the ribosome an RNA-protein complex that catalyzes peptide bond formation revealed that its active site is composed entirely of RNA (Figure 2).
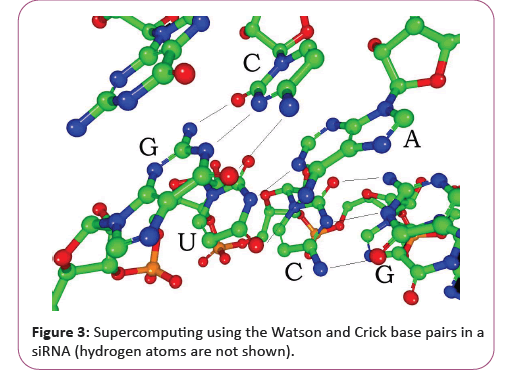
Each nucleotide in RNA contains a ribose sugar, with carbons numbered 1' through 5'. A base is attached to the 1' position, in general, adenine (A), cytosine (C), guanine (G), or uracil (U). Adenine and guanine are purines, cytosine and uracil are pyrimidines. A phosphate group is attached to the 3' position of one ribose and the 5' position of the next. The phosphate groups have a negative charge each, making RNA a charged molecule (polyanion). The bases form hydrogen bonds between cytosine and guanine, between adenine and uracil and between guanine and uracil [16]. However, other interactions are possible, such as a group of adenine bases binding to each other in a bulge, or the GNRA tetra loop that has a guanine–adenine base-pair. An important structural component of RNA that distinguishes it from DNA is the presence of a hydroxyl group at the 2' position of the ribose sugar. The presence of this functional group causes the helix to mostly take the A-form geometry, although in single strand dinucleotide contexts, RNA can rarely also adopt the B-form most commonly observed in DNA. The A- form geometry results in a very deep and narrow major groove and a shallow and wide minor groove. A second consequence of the presence of the 2'-hydroxyl group is that in conformationally flexible regions of an RNA molecule (that is not involved in formation of a double helix), it can chemically attack the adjacent phosphodiester bond to cleave the backbone [17].
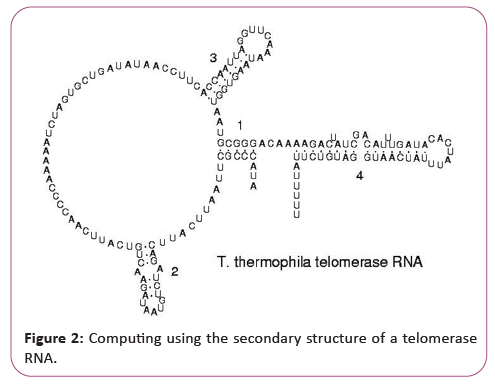
New challenges of supercomputing due to secondary structure of a telomerase RNA
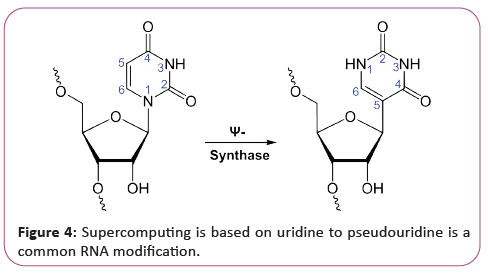
RNA is transcribed with only four bases (adenine, cytosine, guanine and uracil), but these bases and attached sugars can be modified in numerous ways as the RNAs mature [18]. Pseudouridine (Ψ), in which the linkage between uracil and ribose is changed from a C-N bond to a C-C bond, and ribothymidine (T) are found in various places (the most notable ones being in the TΨC loop of tRNA). Another notable modified base is hypoxanthine, a deaminated adenine base whose nucleoside is called inosine (I). Inosine plays a key role in the wobble hypothesis of the genetic code. There are more than 100 other naturally occurring modified nucleosides. The greatest structural diversity of modifications can be found in tRNA, while pseudouridine and nucleosides with 2'-O-methylribose often present in rRNA are the most common. The specific roles of many of these modifications in RNA are not fully understood. However, it is notable that, in ribosomal RNA, many of the post-transcriptional modifications occur in highly functional regions, such as the peptidyl transferase center and the subunit interface, implying that they are important for normal function. The functional form of single-stranded RNA molecules, just like proteins, frequently requires a specific tertiary structure. The scaffold for this structure is provided by secondary structural elements that are hydrogen bonds within the molecule. This leads to several recognizable "domains" of secondary structure like hairpin loops, bulges, and internal loops [19]. Since RNA is charged, metal ions such as Mg2+ are needed to stabilize many secondary and tertiary structures (Figure 3).
The naturally occurring enantiomer of RNA is D-RNA composed of D-ribonucleotides. All chirality centers are located in the D-ribose. By the use of L-ribose or rather L-ribonucleotides, L-RNA can be synthesized. L-RNA is much more stable against degradation by RNAse. Like other structured biopolymers such as proteins, one can define topology of a folded RNA molecule. This is often done based on arrangement of intra-chain contacts within a folded RNA, termed as circuit topology. Synthesis of RNA is usually catalyzed by an enzyme RNA polymerase using DNA as a template, a process known as transcription. Initiation of transcription begins with the binding of the enzyme to a promoter sequence in the DNA (usually found "upstream" of a gene) [20]. The DNA double helix is unwound by the helicase activity of the enzyme. The enzyme then progresses along the template strand in the 3' to 5’ direction, synthesizing a complementary RNA molecule with elongation occurring in the 5' to 3' direction. The DNA sequence also dictates where termination of RNA synthesis will occur. Primary transcript RNAs are often modified by enzymes after transcription. For example, a poly (A) tail and a 5' cap are added to eukaryotic pre-mRNA and introns are removed by the spliceosome. There are also a number of RNA-dependent RNA polymerases that use RNA as their template for synthesis of a new strand of RNA. For instance, a number of RNA viruses (such as poliovirus) use this type of enzyme to replicate their genetic material. Also, RNA-dependent RNA polymerase is part of the RNA interference pathway in many organisms [21].
Biological supercomputing using B Types of RNA
The earliest known regulators of gene expression were proteins known as repressors and activators, regulators with specific short binding sites within enhancer regions near the genes to be regulated. More recently, RNAs have been found to regulate genes as well [22]. There are several kinds of RNA-dependent processes in eukaryotes regulating the expression of genes at various points, such as RNAi repressing genes post-transcriptionally, long noncoding RNAs shutting down blocks of chromatin epigenetically, and enhancer RNAs inducing increased gene expression. In addition to these mechanisms in eukaryotes, both bacteria and archaea have been found to use regulatory RNAs extensively [23]. Bacterial small RNA and the CRISPR system are examples of such prokaryotic regulatory RNA systems. Fire and Mello were awarded the 2006 Nobel Prize in Physiology or Medicine for discovering microRNAs (miRNAs), specific short RNA molecules that can base-pair with mRNAs. Post- transcriptional expression levels of many genes can be controlled by RNA interference, in which miRNAs, specific short RNA molecules, pair with mRNA regions and target them for degradation. This antisense-based process involves steps that first process the RNA so that it can base-pair with a region of its target mRNAs. Once the base pairing occurs, other proteins direct the mRNA to be destroyed by nucleases. Fire and Mello were awarded the 2006 Nobel Prize in Physiology or Medicine for this discovery [24].
Long non-coding RNA and our supercomputer
Next to be linked to regulation were existing and other long noncoding RNAs associated with X chromosome inactivation. Their roles, at first mysterious, were shown by Jeannie T. Lee and others to be the silencing of blocks of chromatin via recruitment of Polycomb complex so that messenger RNA could not be transcribed from them. Additional lncRNAs currently defined as RNAs of more than 200 base pairs that do not appear to have coding potential, have been found associated with regulation of stem cell pluripotency and cell division [25]. The third major group of regulatory RNAs is called enhancer RNAs. It is not clear at present whether they are a unique category of RNAs of various lengths or constitute a distinct subset of lncRNAs. In any case, they are transcribed from enhancers, which are known regulatory sites in the DNA near genes they regulate them up-regulate the transcription of the genes under control of the enhancer from which they are transcribed. This facilitates us to construct this supercomputer. At first, regulatory RNA was thought to be a eukaryotic phenomenon, a part of the explanation for why so much more transcription in higher organisms was seen than had been predicted. But as soon as researchers began to look for possible RNA regulators in bacteria, they turned up there as well, termed as small RNA (sRNA) [26]. Currently, the ubiquitous nature of systems of RNA regulation of genes has been discussed as support for the RNA World theory. Bacterial small RNAs generally act via antisense pairing with mRNA to down- regulate its translation, either by affecting stability or affecting cis-binding ability. Riboswitches have also been discovered. They are cisacting regulatory RNA sequences acting allosterically. They change shape when they bind metabolites so that they gain or lose the ability to bind chromatin to regulate expression of genes. All these things are very good for our supercomputer, archaea also have systems of regulatory RNA [27]. The CRISPR system recently being used to edit DNA in situ, acts via regulatory RNAs in archaea and bacteria to provide protection against virus invaders. Many RNAs are involved in modifying other RNAs. Introns are spliced out of pre-mRNA by spliceosomes, which contain several small nuclear RNAs (snRNA), or the introns can be ribozymes that are spliced by them. RNA can also be altered by having its nucleotides modified to nucleotides other than A, C, G and U. In eukaryotes, modifications of RNA nucleotides are in general directed by small nuclear RNAs (snoRNA; 60-300 nt), found in the nucleolus and cajal bodies [28]. snoRNAs associate with enzymes and guide them to a spot on an RNA by base pairing to that RNA. These enzymes then perform the nucleotide modification. rRNAs and tRNAs are extensively modified, but snRNAs and mRNAs can also be the target of base modification. RNA can also be methylated (Figure 4).
Why our supercomputer finally can be constructed: Like DNA, RNA can carry genetic information. RNA viruses have genomes composed of RNA that encodes a number of proteins [29]. The viral genome is replicated by some of those proteins, while other proteins protect the genome as the virus particle moves to a new host cell. Viroid’s are another group of pathogens, but they consist only of RNA, do not encode any protein and are replicated by a host plant cells polymerase.
Reverse transcribing viruses replicate their genomes by reverse transcribing DNA copies from their RNA; these DNA copies are then transcribed to new RNA [30]. Retro transposons also spread by copying DNA and RNA from one another, and telomerase contains an RNA that is used as template for building the ends of eukaryotic chromosomes. Double-stranded RNA (dsRNA) is RNA with two complementary strands, similar to the DNA found in all cells, but with the replacement of thymine by uracil. dsRNA forms the genetic material of some viruses (double- stranded RNA viruses). Double-stranded RNA, such as viral RNA or siRNA, can trigger RNA interference in eukaryotes, as well as interferon response in vertebrates [31].
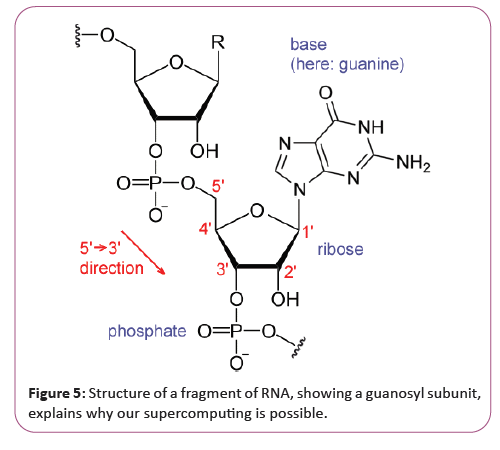
In the late 1970s, it was shown that there is a single stranded covalently closed, i.e. circular form of RNA expressed throughout the animal and plant kingdom (see circular RNA). Circular RNAs are thought to arise via a "back-splice" reaction where the spliceosome joins a downstream donor to an upstream acceptor splice site. So far the function of circular RNAs is largely unknown, although for few examples a microRNA sponging activity has been demonstrated. Why this supercomputer can be constructed [32]. Our Supercomputer is possible because the Research on RNA has led to many important biological discoveries and numerous Nobel Prizes. Nucleic acids were discovered in 1868 by Friedrich Miescher, who called the material 'nuclein' since it was found in the nucleus. It was later discovered that prokaryotic cells, which do not have a nucleus, also contain nucleic acids. The role of RNA in protein synthesis was suspected already in 1939. Severo Ochoa won the 1959 Nobel Prize in Medicine (shared with Arthur Kornberg) after he discovered an enzyme that can synthesize RNA in the laboratory. However, the enzyme discovered by Ochoa (polynucleotide phosphorylase) was later shown to be responsible for RNA degradation, not RNA synthesis. In hybridized two separate strands of RNA to form the first crystal of RNA whose structure could be determined by X-ray crystallography [33]. The sequence of the 77 nucleotides of yeast tRNA was found (Figure 5).
In the early 1970s, retroviruses and reverse transcriptase were discovered, showing for the first time that enzymes could copy RNA into DNA (the opposite of the usual route for transmission of genetic information) [34]. For this work, determined the first complete nucleotide sequence of an RNA virus genome, that of bacteriophage MS2. In 1977, introns and RNA splicing were discovered in both mammalian viruses and in cellular genes, resulting in Nobel to Philip Sharp and Richard Roberts. Catalytic RNA molecules (ribozymes) were discovered in the early. In 1990, it was found in Petunia that introduced genes can silence similar genes of the plant's own, now known to be a result of RNA interference [35]. At about the same time, 22 long RNAs, now called microRNAs, were found to have a role in the development of C elegans (Figure 6).
Results
Studies on RNA interference gleaned a Nobel Prize for Andrew Fire and Craig Mello in 2006, and another Nobel was awarded for studies on the transcription of RNA to Roger Kornberg in the same year. The discovery of gene regulatory RNAs has led to attempts to develop drugs made of RNA, such as siRNA, to silence genes. Adding to the Nobel prizes awarded for research on RNA in 2009 it was awarded for the elucidation of the atomic structure of the ribosome to Venki Ramakrishnan, Tom Steitz, and Ada Yonath In 1967, Carl Woese hypothesized that RNA might be catalytic and suggested that the earliest forms of life (self-replicating molecules) could have relied on RNA both to carry genetic information and to catalyze biochemical reactions an RNA world. In March 2020, complex DNA and RNA nucleotides, including uracil, cytosine and thymine, were reportedly formed in the laboratory under outer space conditions, using starter chemicals, such as pyrimidine, an organic compound commonly found in meteorites. Pyrimidine, like polycyclic aromatic hydrocarbons (PAHs), is one of the most carbon-rich compounds found in the Universe and may have been formed in red giants or in interstellar dust and gas clouds.
Discussion and Conclusion
In this paper, we proposed a biological supercomputer which is based in RNA molecules Biological computers are made from living cells. Instead of electrical wiring and signaling, biological computers use chemical inputs and other biologically derived molecules such as proteins and DNA. In this paper we constructed a biological supercomputer which is based in RNA molecules using Quantum Mechanics and Nanoptics. We used simulation with Neural Networks and Fuzzy Logic. The theoretical results are supported by numerical analysis and two experiments.
References
- Badieyan ZS, Aneja MK, Plank C. (2015) Magnetofection: A Versatile Approach for Messenger RNA Delivery. Mol Thera 23: 143-144.
- Shukla RN. (2014) Analysis of Chromosomes.
- Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry (5th ed.) WH Freeman and Company 118–119.
- Tinoco J. Bustamante C (1999) How RNA folds. J Mol Biol 293: 271-281.
- Higgs PG (2000) RNA secondary structure physical and computational aspects. Q Rev Biophys 33: 199-253.
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. J Sci 289: 920-930.
- Barciszewski J, Frederic B, Clark C (1999) RNA biochemistry and biotechnology. Springer 23: 73-87.
- Salazar M, Fedoroff OY, Miller JM, Ribeiro NS, Reid BR (1993) The DNA strand in DNA and RNA hybrid duplexes is neither B-form nor A-form in solution. Anal Biochem 32: 4207-4215.
- Yu Q, Morrow CD (2001) Identification of critical elements in the tRNA acceptor stem and TΨC loop necessary for human immunodeficiency virus type 1 infectivity. J Virol 75: 4902-4906.
- Sedova A, Banavali NK (2016) RNA approaches the B‐form in stacked single strand dinucleotide contexts. J Biopoly 105: 65-82.
- Hermann T, Patel DJ (2000) RNA bulges as architectural and recognition motifs. J Structure 8: 47-54.
- Mikkola S, Stenman E, Nurmi K, Yousefi-Salakdeh E, Strömberg R. (1999) The mechanism of the metal ion promoted cleavage of RNA phosphodiester bonds involves a general acid catalysis by the metal aquo ion on the departure of the leaving group. J Chem Soc 2: 1619-1626.
- Jankowski JA, Polak JM (1996) Clinical gene analysis and manipulation: Tools, techniques and troubleshooting. Cambridge University Press.
- Elliott MS, Trewyn RW (1984) Inosine biosynthesis in transfer RNA by an enzymatic insertion of hypoxanthine. J Biol Chem 259: 2407-2410.
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA et al. (2010) The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 10: 195-201.
- Söll D, Soll D, RajBhandary U, RajBhandary TL (1995) Editors. tRNA: structure, biosynthesis, and function.
- Jády BE, Kiss T (2001) A small nucleolar guide RNA functions both in 2′‐O‐ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J 20: 541-551.
- King TH, Liu B, McCully RR, Fournier MJ (2003) Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 11: 425-435.
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci 101: 7287-7292.
- Tan ZJ, Chen SJ (2008) Salt dependence of nucleic acid hairpin stability. Biophys. J 95: 738-752.
- Vater A, Klussmann S (2015) Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics. Drug Discov Today 20: 147-155.
- Hansen JL, Long AM, Schultz SC (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. J Structure 5: 1109-1122.
- Ahlquist P (2002) RNA-dependent RNA polymerases, viruses, and RNA silencing. J Science 296: 1270-1273.
- Lee JC, Gutell RR (2000) Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J Mol Biol 344: 1225-1249.
- Cooper GC, Hausman RE (2004) The Cell: A Molecular Approach 3rd Edition 339–344.
- Mattick JS, Gagen MJ (2001) The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol 18: 1611-1630.
- Mattick JS (2001) Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep 2: 986-991.
- Mattick JS (2003) Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. J Bioessays 25: 930-939.
- Mattick JS (2004) The hidden genetic program of complex organisms. Sci Am 291: 60-67.
- Kossen K, Vaish NK, Jadhav VR, Pasko C, Wang H et al. (2004) High-throughput ribozyme-based assays for detection of viral nucleic acids. Chem Bio 11: 807-815.
- Storz G (2002) An expanding universe of noncoding RNAs. J Science 296: 1260-1263.
- Fatica A, Bozzoni I (2004) Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet 15: 7-21.
- Chen Q, Yan M, Cao Z, Li X, Zhang Y et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. J Science 351: 397-400.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences